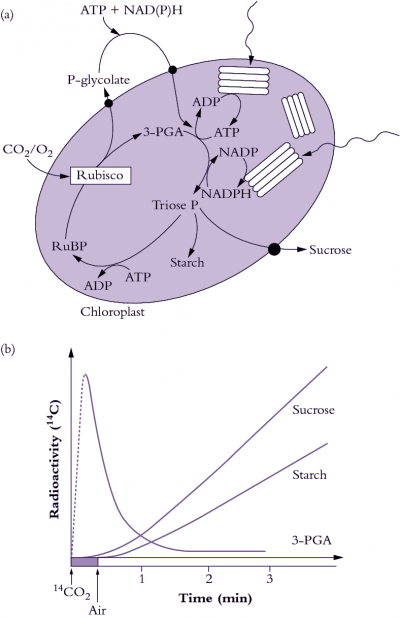
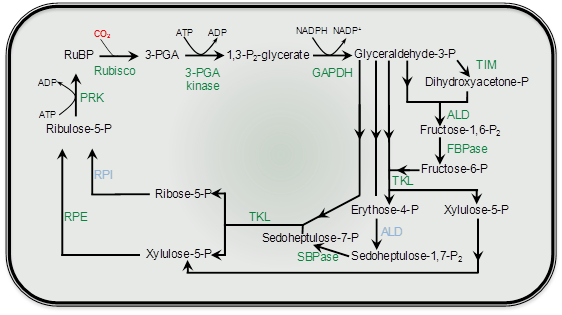
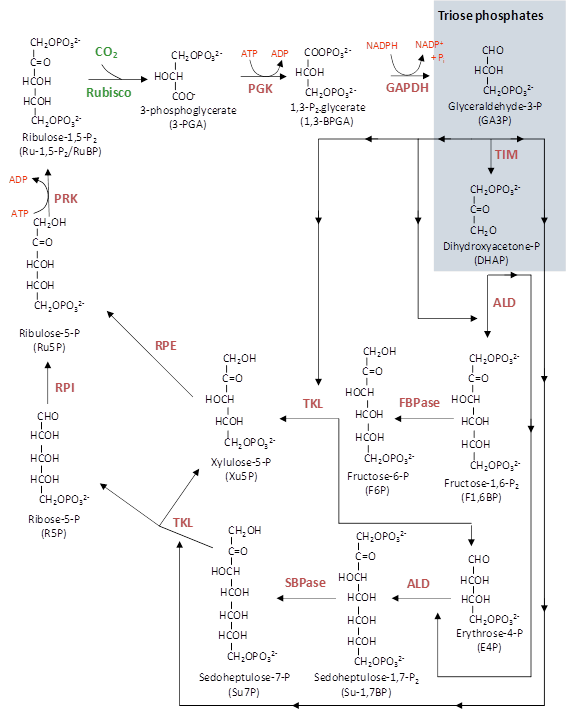
Photosynthetic carbon fixation in air is constrained by the kinetic properties of Rubisco. Form I Rubisco in higher plants is a large protein (approximately 550 kDa) comprised of eight large (approx. 50-55 kDa) and eight small subunits (approx. 13-18 kDa) to form an L8S8 hexadecamer. Rubisco synthesis and assembly in higher plants is a complex process whereby the large subunit gene (rbcL) is encoded in the chloroplast genome, while the small subunit genes (rbcS) are encoded as a multi-gene family in the nucleus. The Rubisco small subunits are translated as precursors in thc cytosol and are equipped with a transit-peptide to target them to the chloroplast. Upon import in the chloroplast the transit-peptide is cleaved by a stromal peptidase and the N-terminus modified by methylation of the n-terminal methionine. The large subunits are synthesised within the chloroplast and also post-translationally modified through the removal of the the N-terminal methionine and serine amino acids and the subsequent acetylation of proline at the N-terminus and the methylation of lysine at position 14. The assembly of large and small subunits into functional hexadecameric Rubisco is reliant on the coordination of chloroplast-localised chaperones.
Despite selection pressure over evolutionary history, Rubisco remains an inefficient catalyst (Spreitzer and Salvucci 2002). Therefore, to achieve a productive maximum CO2 assimilation rate (Amax), plants must compensate for catalytic inefficiency by investing large amounts of nitrogen in Rubisco. Consequently, Rubisco comprises more than 50% of leaf soluble protein in C3 plants. On a global scale, this investment equates to around 10 kg of nitrogen per person!
More than 1000 million years of evolution has still not resulted in a ‘better’ Rubisco adapted for the current and future concentrations of CO2. Such a highly conserved catalytic protein is an outcome of thermodynamic and mechanistic difficulties inherent to this reaction. Rubisco requires carbamylation of the absolutely-conserved residue K201 that is then stabilised by the binding of Mg2+. Without this activation step Rubisco is unable to function. The fixation of CO2 to RuBP to form two molecules of 3-PGA is a five step catalytic process that produces highly reactive transition state intermediates that bind CO2. The highly reactive transition states make Rubisco prone to generating misfiring products, which generate inhibitors within the active site. Therefore, Rubisco requires its own catalytic protection enzyme Rubisco activase. Plants devoid of this enzyme fail to grow properly in air as the activation and subsequent activity of Rubisco is impeded (Portis and Salvucci 2002). Rubisco activase is an ATP-dependent process that removes inhibitors from the active site of Rubisco allowing for activation and catalysis to proceed. Recently, the crystal structure of Rubisco activase has been solved, which will provide key insight into the molecular interaction between Rubisco and Rubisco activase (reviewed by Portis et al. 2008).
Rubisco first evolved when the earth’s atmosphere was rich in CO2, but virtually devoid of O2. With the advent of oxygen-producing photosynthesis by land plants, and the resulting increases in atmospheric O2, one key deficiency of this enzyme became apparent. Rubisco would not only catalyse fixation of CO2 but would also permit incorporation of O2 into RuBP to produce, instead of two molecules of 3-PGA, just one molecule of 3-PGA with one molecule of a two-carbon compound, 2-phosphoglycolate (Section 2.3). Indeed, CO2 and O2 compete directly for access to the active sites of Rubisco. So feeble is Rubisco’s ability to distinguish between these two substrates that in air (20% O2) approximately one molecule of O2 is fixed for every three molecules of CO2.
Fixation of O2 and subsequent photorespiration (Section 2.3) is an energy-consuming process, due to competition between O2 and CO2 for RuBP, plus the energy cost of converting the phosphoglycolate product to a form which can be recycled in the PCR cycle. This energy cost is increased at higher temperatures because O2 competes more effectively with CO2 at the active site of Rubisco. Such sensitivity to temperature × O2 explains why CO2 enrichment, which reduces photorespiration, has a proportionally larger effect upon net carbon gain at higher temperatures than at lower temperatures (Section 13.3).
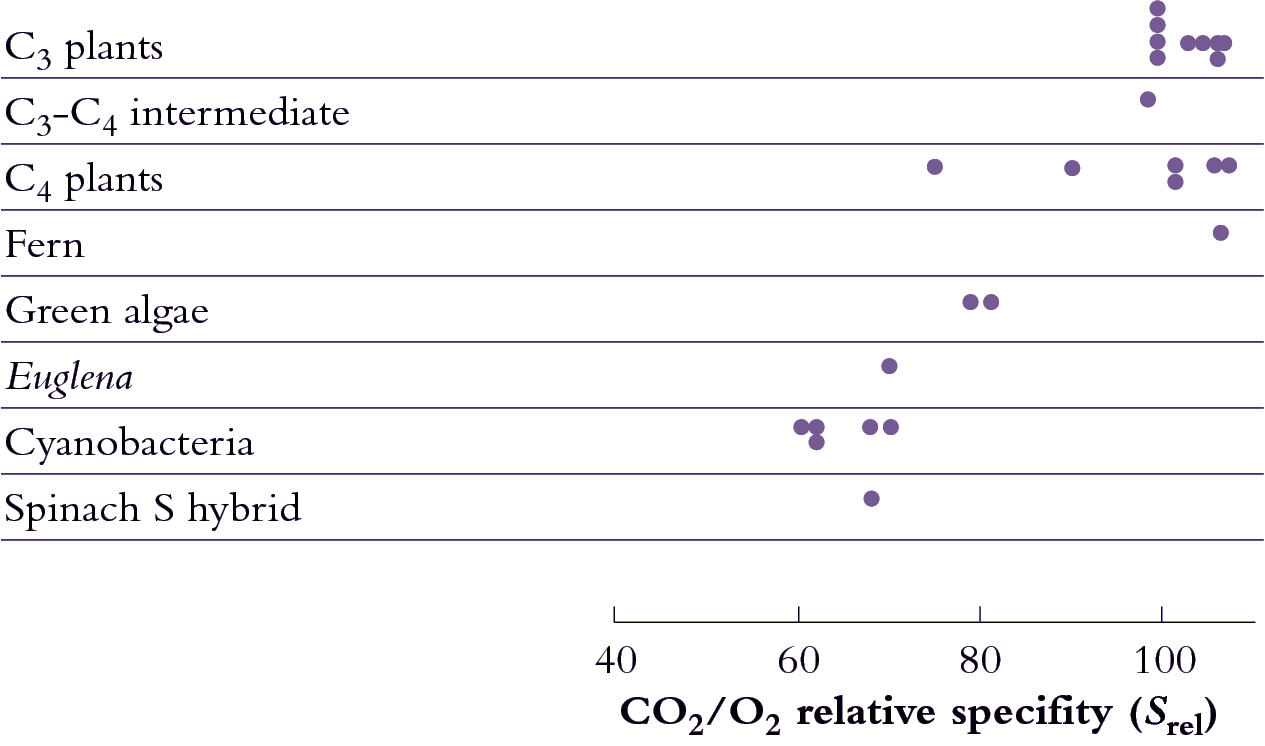
Notwithstanding a meagre catalytic effectiveness in present day Rubisco, more efficient variants would still have had a selective advantage, and especially during those times in the earth’s geological history when atmospheric CO2 concentration was decreasing. Indeed there has been some improvement (Figure 2.3) such that specificity towards CO2 as opposed to O2 has improved significantly. Recently evolved angiosperms show a relative specificity almost twice that of 'older' organisms such as photosynthetic bacteria.
Despite such improvement, Rubisco remains seemingly maladapted to its cardinal role in global carbon uptake, and in response to selection pressure for more efficient variants of CO2 assimilation, vascular plants have evolved with photosynthetic mechanisms that alleviate an inefficient Rubisco. One key feature of such devices is a mechanism to increase CO2 concentration at active sites within photosynthetic tissues. Some of these photosynthetic pathways are dealt with below.