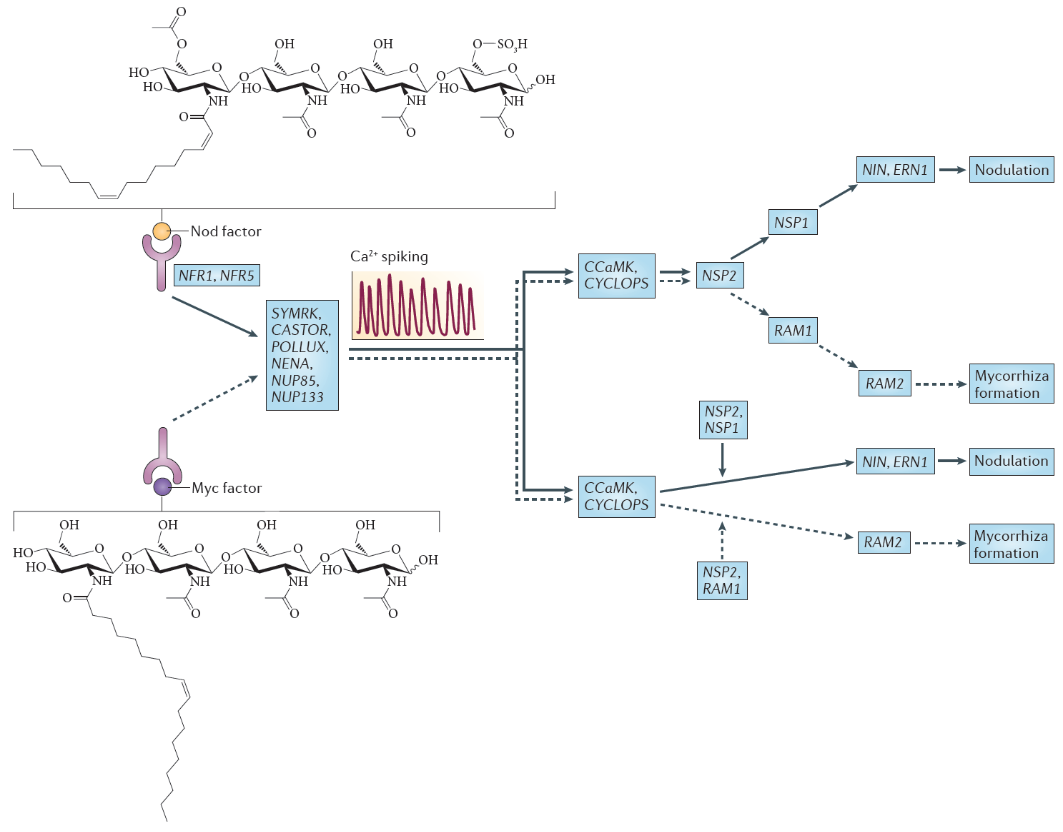
Figure 4.45 Nod factor and Myc factor signalling. The structure of the predominant Nod factor from Sinorhizobium meliloti, the symbiont of the legume Medicago truncatula, is shown (top), in comparison with the structure of the non-sulphated lipochitooligosaccharide Myc factor produced by the arbuscular mycorrhizal fungus Glomus intraradices (bottom). These molecules induce the symbiosis signalling pathway, that shares a calcium spiking response. There are two different interpretations for the genetic overview of this signalling pathway. These interpretations are an amalgamation of genetic analyses in Lotus japonicus and Medicago truncatula; gene names are indicated for the model legume L. japonicus. In the first interpretation (top), the symbiosis signalling pathway transmits the signal through the transcription factors encoded by nodulation signalling pathway 2 (NSP2), NSP1and RAM1 (required for arbuscular mycorrhization 1), whereas in the second interpretation (bottom), symbiosis signalling occurs in parallel to the signalling through these GRAS domain transcription factors. Names for these genes in M. truncatula are as follows: Nod factor receptor 1 (NFR1) is LysM domain receptor kinase 3 (LYK3); NFR5 is Nod factor perception (NFP); symbiosis receptor-like kinase (SYMRK) is DMI2; POLLUX is DMI1; calcium- and calmodulin-dependent serine/threonine protein kinase (CCAMK) is also known as DMI3; CYCLOPS is interacting protein of DMI3 (IPD3). (Reproduced by permission from Macmillan Publishers Ltd from G.E.D. Oldroyd, Nature Rev Microbiol 11: 252-632, 2013)