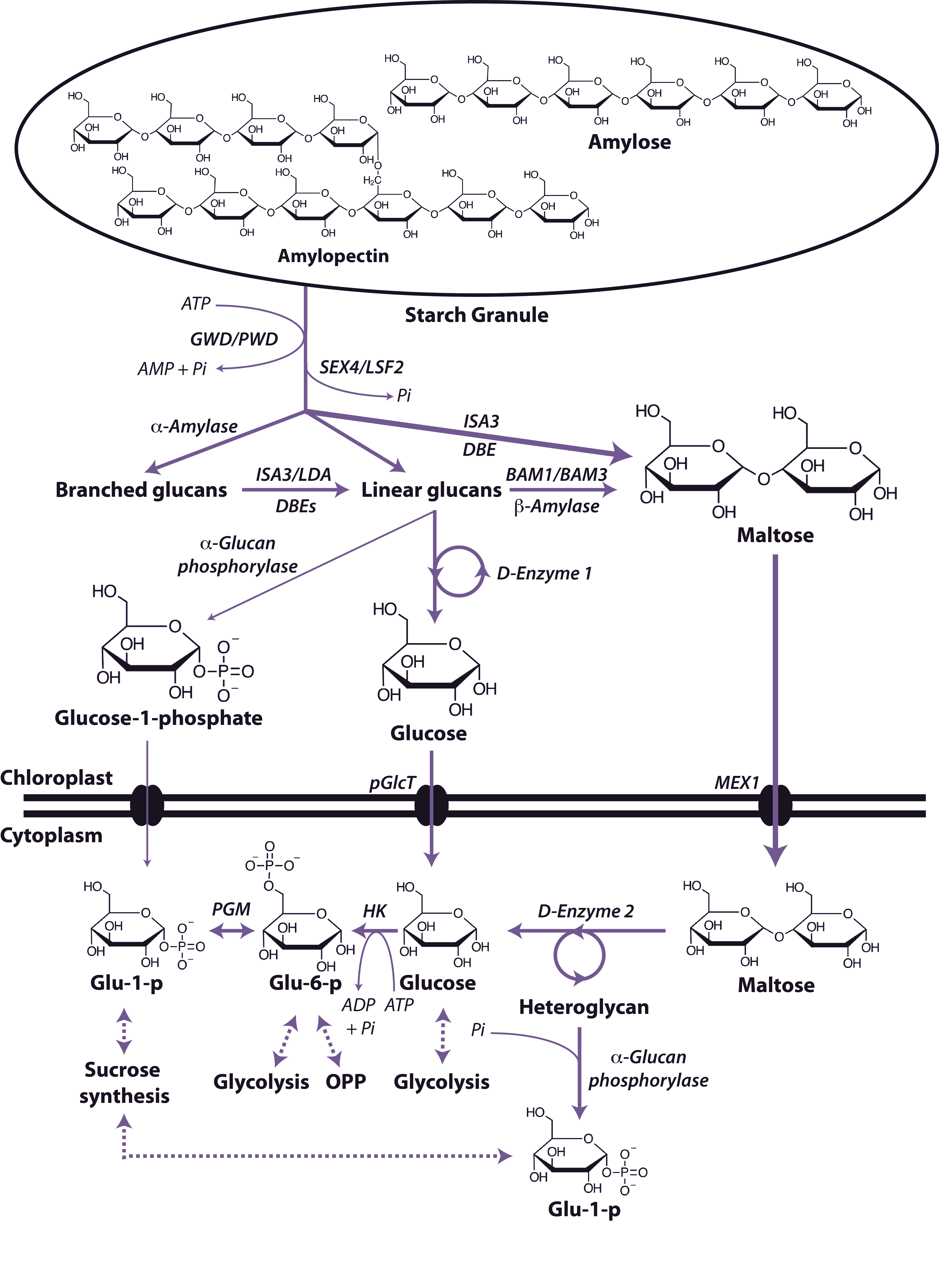
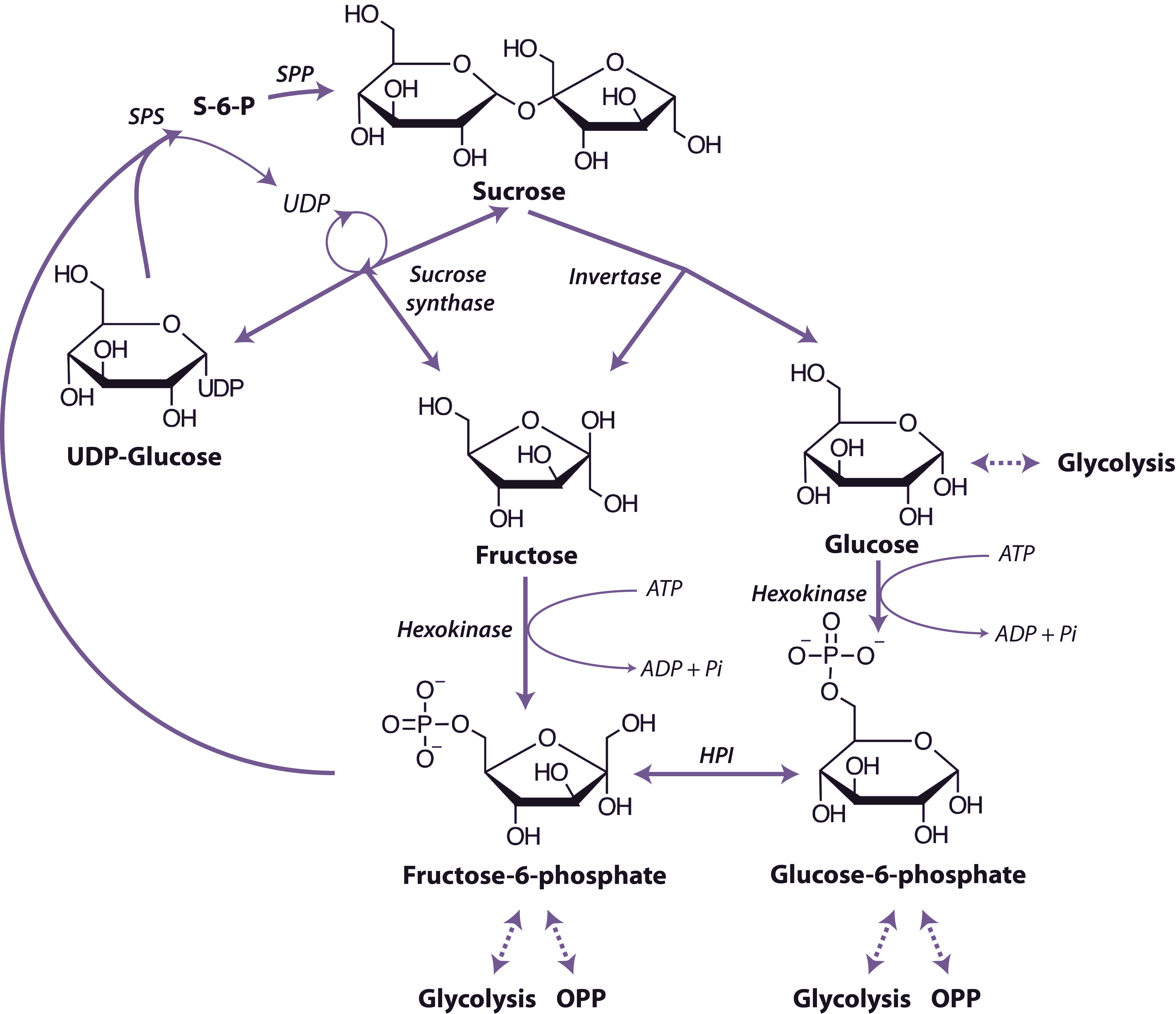
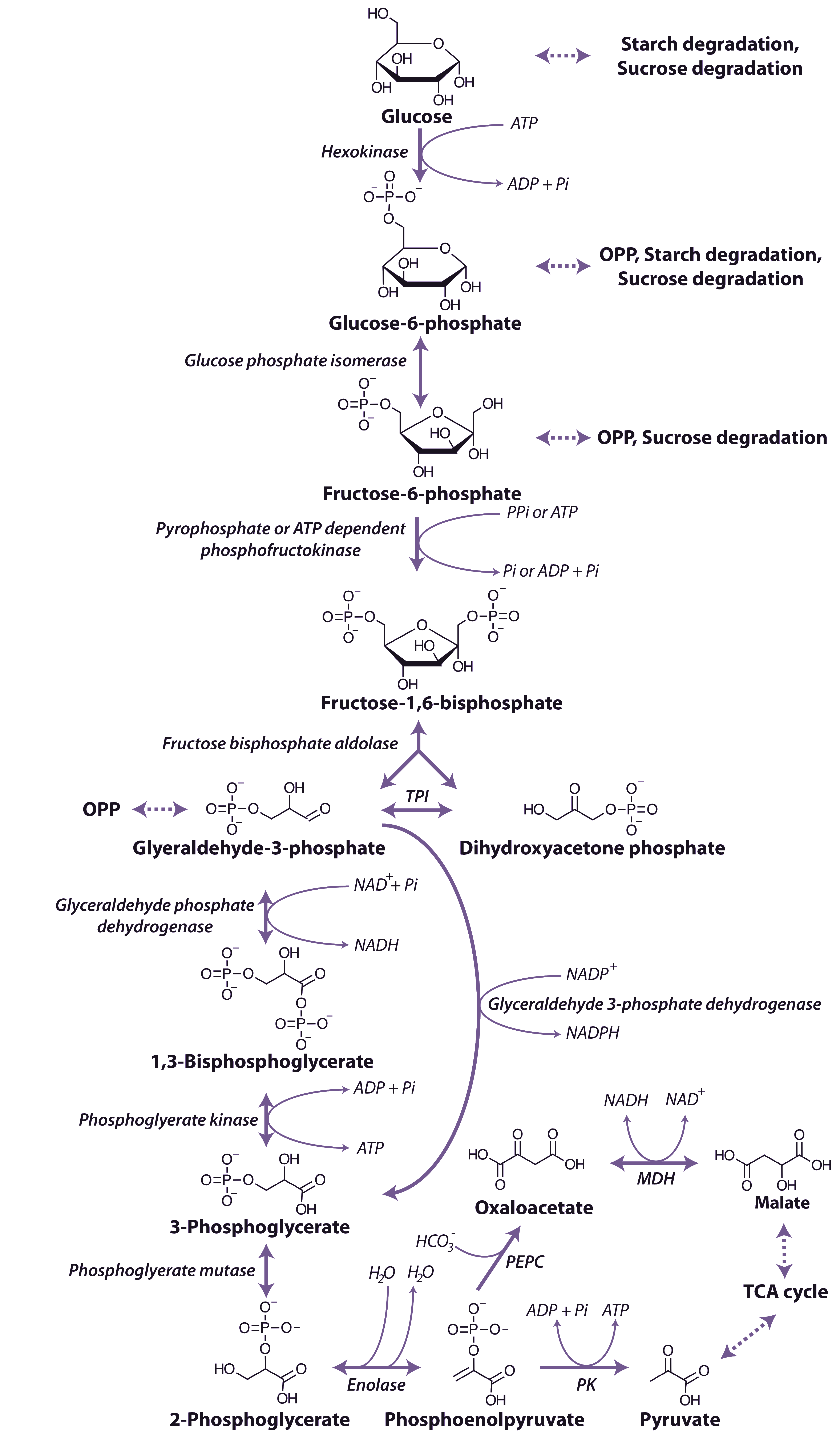
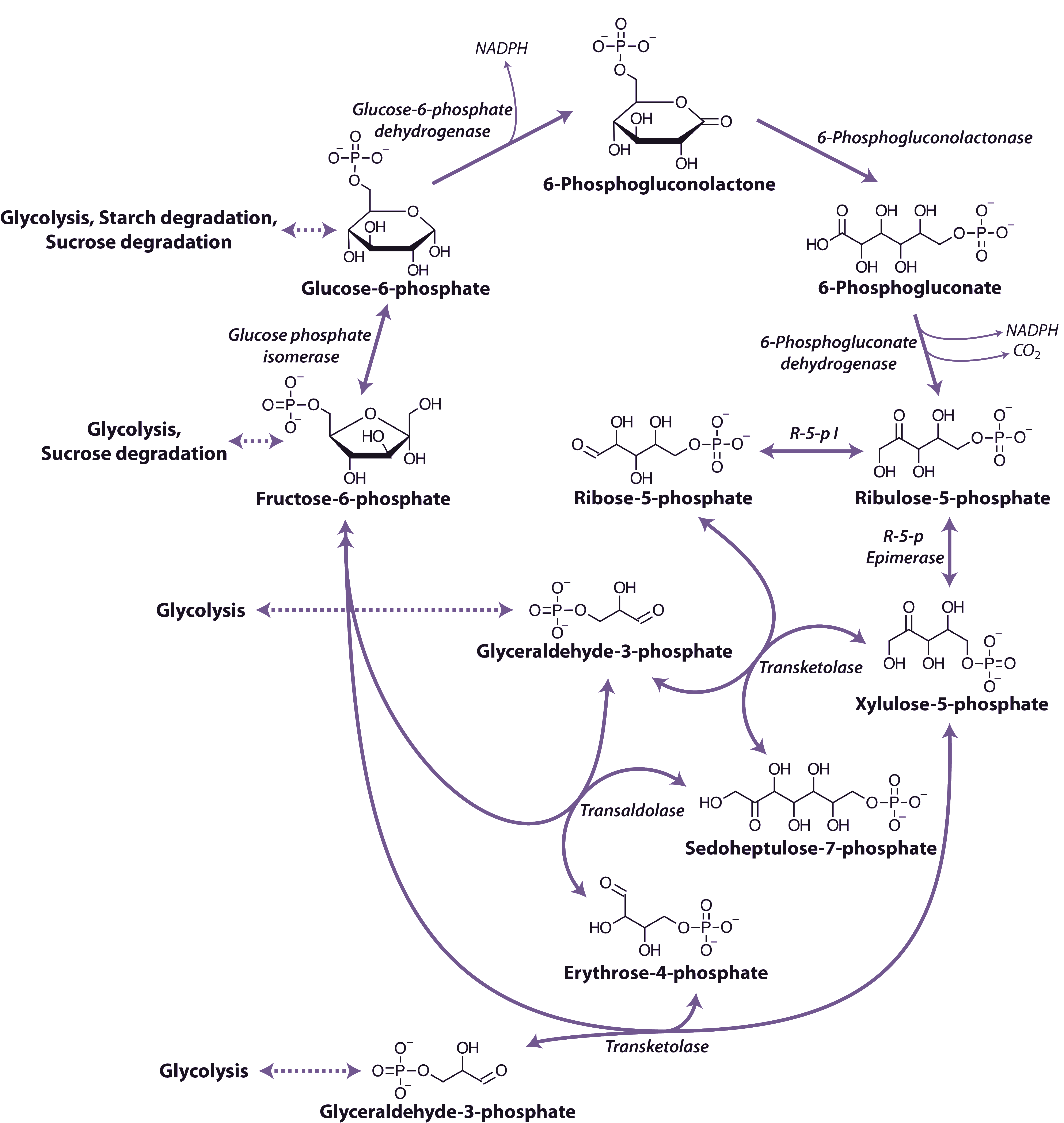
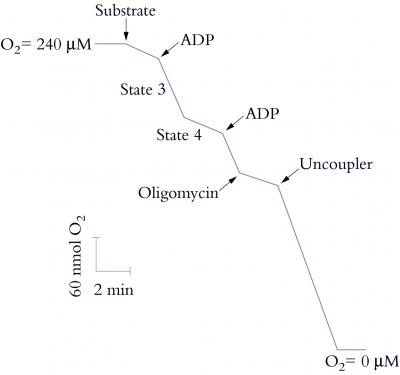
Organic acids such as pyruvate and malate produced in the cytosol by processes described above are further oxidised in mitochondria by the tricarboxylic acid (TCA) cycle and subsequent respiratory chain. Energy released by this oxidation is used to synthesise ATP which is then exported to the cytosol for use in biosynthesis and growth.
(a) Mitochondrial structure
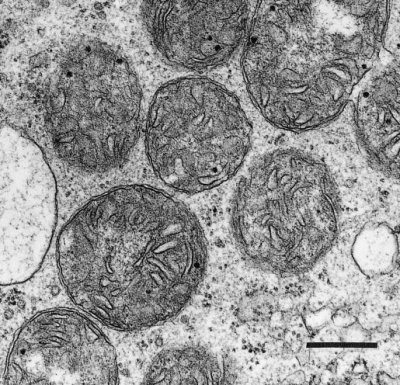
Plant mitochondria (Figure 2.23) are typically double-membrane organelles where the inner membrane is invaginated to form folds known as cristae to increase the surface area of the membrane. The outer membrane contains relatively few proteins (<100) and is permeable to most small compounds (< Mr=5 kDa) due to the presence of the pore-forming protein VDAC (voltage dependent anion channel) which is a member of the porin family of ion channels. The inner membrane is the main permeability barrier of the organelle and controls the movement of molecules by means of a series of carrier proteins many of which are members of mitochondrial substrate carrier family (MSCF). The inner membrane also houses the large complexes that carry out oxidative phosphorylation and encloses the soluble matrix which contains the enzymes of the TCA cycle and many other soluble proteins involved in a myriad of mitochondrial functions.
Mitochondria are semi-autonomous organelles with their own DNA and protein synthesis machinery. However, the mitochondrial genome encodes only a small portion of the proteins which make up the mitochondrion; the rest are encoded on nuclear genes and synthesised in the cytosol. These proteins are the transported into the mitochondrion by the protein import machinery and assembled with the mitochondrially synthesised subunits to form the large respiratory complexes. The number of mitochondria per cell varies with tissue type (from a few hundred in mature differentiated tissue to some thousands in specialised cells). Understandably, more active cells, with high energy demands, such as those in growing meristems are generally equipped with larger numbers of mitochondria per unit cell volume, and consequently show faster respiration rates.
(b) Mitochondrial substrates
Two substrates are produced from glycolytic PEP for oxidation in mitochondria: malate and pyruvate (Figure 2.21). These compounds are thought to be the most abundant mitochondrial substrates in vivo. However, amino acids may also serve as substrates for mitochondrial respiration in some tissues, particularly in seeds rich in stored protein or under conditions of sugar depletion such as extended darkness, shading and senescence. β-oxidation of fatty acids typically does not occur in plant mitochondria, this oxidation is principally carried out in peroxisomes in plants.
(c) Carbon metabolism in mitochondria
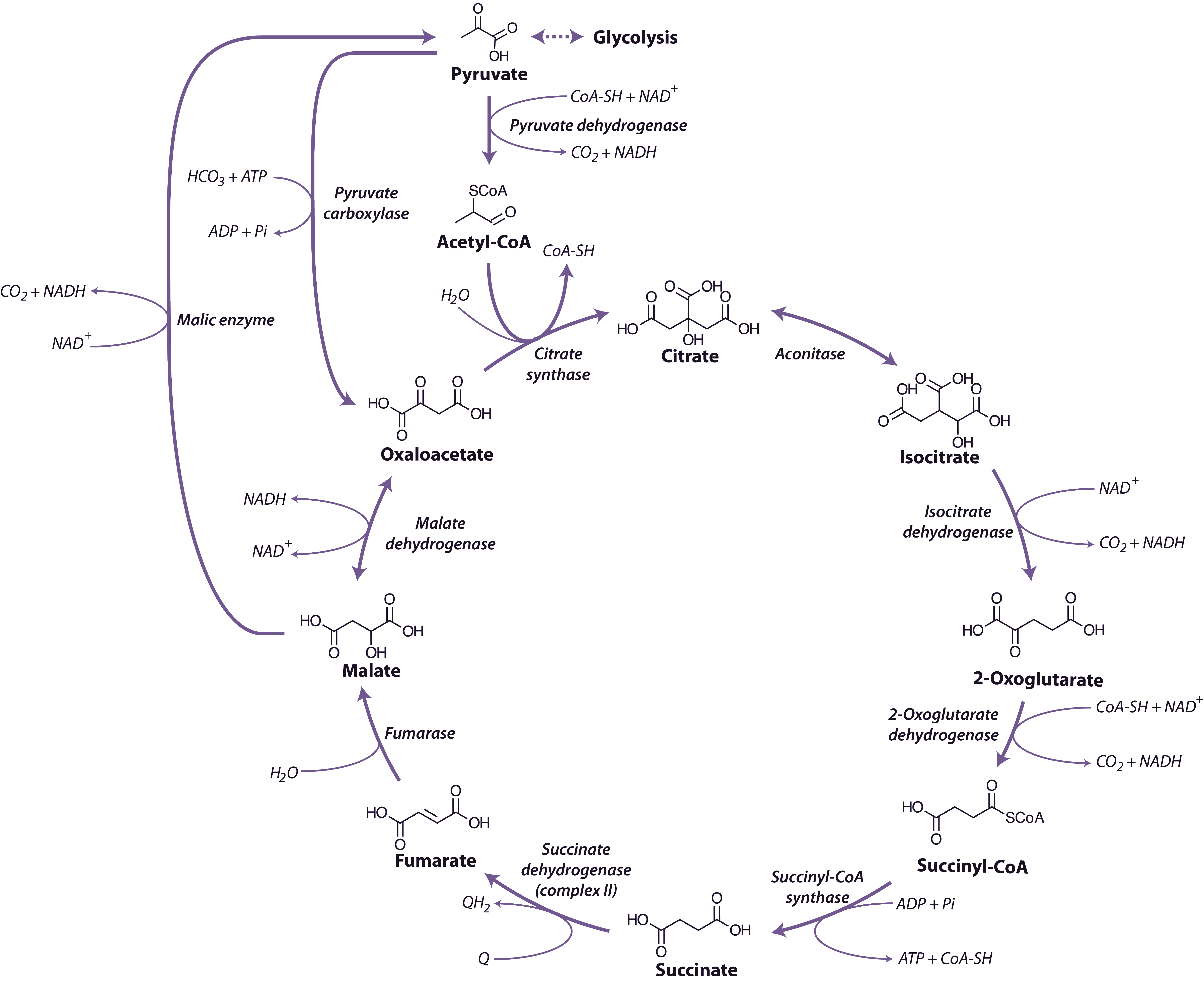
Malate and pyruvate enter the mitochondrial matrix across the inner membrane via separate carriers. Malate is then oxidised by either malate dehydrogenase (a separate enzyme isoform from that in the cytosol), which yields oxaloacetate (OAA) and reduced nicotinamide adenine dinucleotide (NADH), or NAD+-linked malic enzyme, which yields pyruvate and NADH and releases CO2 (Figure 2.24). Cytosolic pyruvate carboxylase is an alternative means of providing substrate to mitochondria by combining pyruvate with HCO3 to yield OAA that can then be imported into mitochondria.
Pyruvate formed either from malate and malic enzyme or transported directly from the cytosol is oxidised inside mitochondria by the pyruvate dehydrogenase complex (PDC) to form CO2, acetyl-CoA and NADH. This enzyme, which requires coenzyme A, thiamine pyrophosphate and lipoic acid as cofactors, effectively links the TCA cycle to glycolysis. PDC comprises three enzymes E1 (2-oxo acid dehydrogenase), E2 (acyltransferase) and E3 (lipoamide dehydrogenase). This complex is regulated by phosphorylation of the E1 subunit, lowering PDC activity in the day and increasing PDC activity at night. Pyruvate dehydrogenase is also subject to feedback inhibition from acetyl-CoA and NADH.
(d) Tricarboxylic acid (TCA) cycle
The TCA cycle begins with the condensation of acetyl-CoA and OAA, to form the six-carbon molecule citrate and the release coenzyme A (CoA) (Figure 2.24) in a reaction catalysed by citrate synthase. Aconitase catalyses the next step, converting citrate to isocitrate in a two-step reaction (dehydration/hydration) with cis-aconitate as an intermediate.
NAD-linked isocitrate dehydrogenase then oxidatively decarboxylates isocitrate to form CO2 and 2-oxoglutarate, and reduce NAD+ to NADH. The 2-oxoglutarate formed is also oxidatively decarboxylated to succinyl-CoA in a reaction catalysed by the enzyme 2-oxoglutarate dehydrogenase. This enzyme complex has similarities to pyruvate dehydrogenase and its reaction is analogous to the formation of acetyl-CoA from pyruvate by pyruvate dehydrogenase. The reaction mechanisms are also very similar, with 3 subunit enzymes, but 2-oxoglutarate dehydrogenase is not subject to the phosphorylation control that regulates pyruvate dehydrogenase. Succinyl-CoA synthase then catalyses the conversion of succinyl-CoA to succinate, with the concomitant phosphorylation of ADP to ATP, the only substrate-level phosphorylation step in the mitochondrion. This enzyme in plants differs from its mammalian counterpart in that it is specific for ADP rather than GDP.
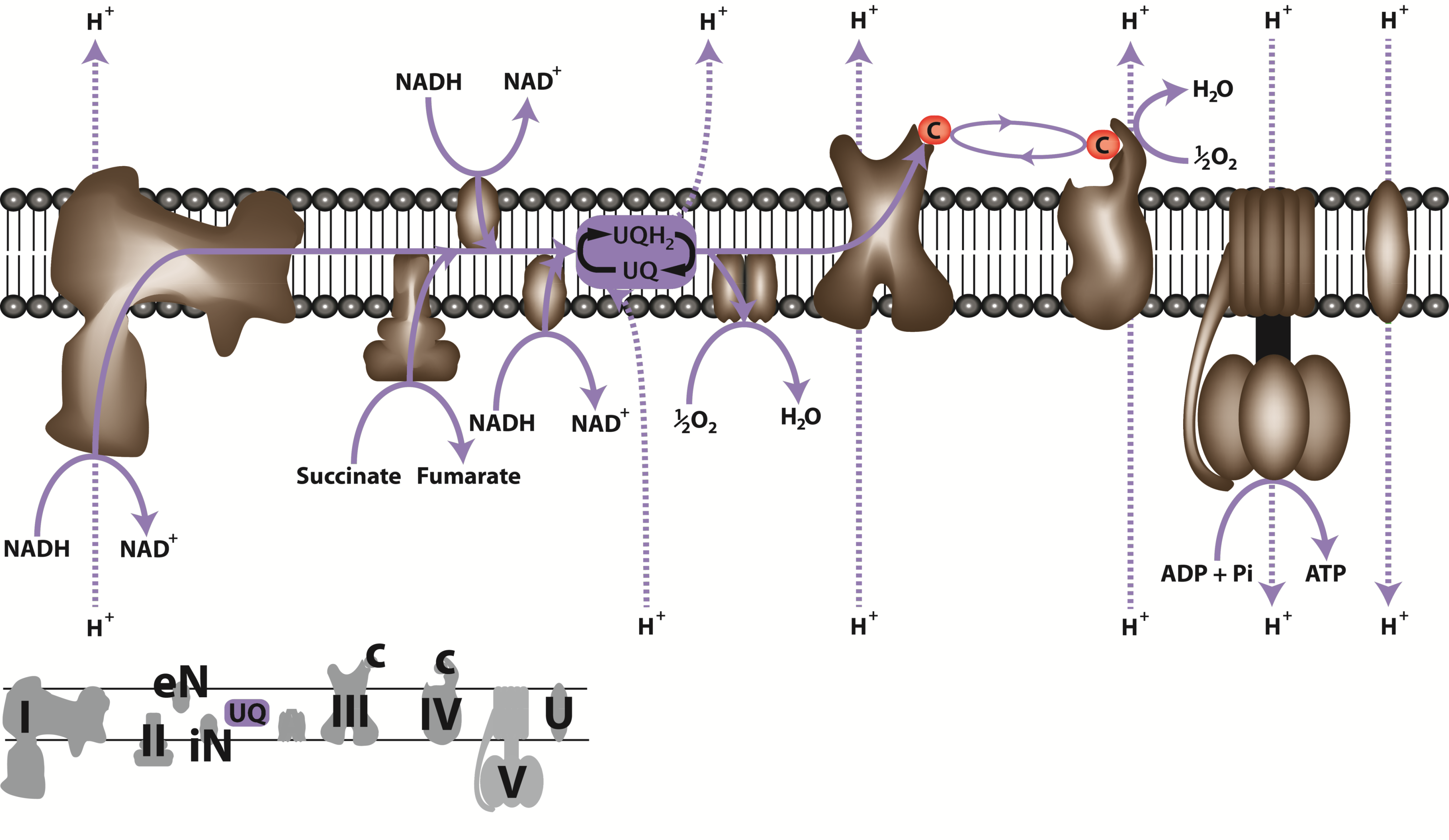
Succinate dehydrogenase (SDH, Complex II), which catalyses the oxidation of succinate to fumarate, is the only membrane-bound enzyme of the TCA cycle and is part of the respiratory electron transfer chain (Figure 2.25). SDH is a large complex consisting of four core subunits, as well as number other associated subunits.
Fumarase catalyses the hydration of fumarate to malate followed by malate dehydrogenase that catalyses the final step of the TCA cycle, oxidising malate to OAA and producing NADH. The reaction is freely reversible, although the equilibrium constant strongly favours the reduction of OAA, necessitating rapid turnover of OAA and NADH to maintain this reaction in a forward direction.
Overall, during one turn of the cycle, three carbons of pyruvate are released as CO2, one molecule of ATP is formed directly, and four NADH and one FADH2 are produced. The strong reductants are oxidised in the respiratory chain to reduce O2 and produce ATP. Although most of the TCA cycle enzymes in plant mitochondria are NAD linked, NADP-dependent isoforms of isocitrate and malate dehydrogenases also exist, and these may play a role in a protective reductive cycle in the matrix.
Regulation of carbon flux through the TCA cycle probably occurs via phosphorylation/dephosphorylation of pyruvate dehydrogenase, which will depend in turn on mitochondrial energy status and feedback inhibition of various enzymes by NADH and acetyl-CoA. The rate of cycle turnover thus depends on the rate of electron flow through the respiratory chain (to reoxidise NADH) and the utilisation of ATP in the cell to provide ADP for substrate level and oxidative phoshorylation. TCA cycle turnover will also depend on the rate of substrate provision by reactions in chloroplasts and cytosol. A number of studies of TCA cycle mutants have demonstrated the wide impact these enzymes have not simply on TCA cycle function but as steps for the delivery of organic acids for other processes in plant cells such a photosynthetic performance, plant biomass, root growth, photorespiration, nitrogen assimilation, amino acid metabolism, and stomatal function.