Different nutrient acquisition strategies. (figure by S. Buckley)
Chapter editors: Rana Munns and Susanne Schmidt
Contributing Authors: MC Brundrett1,2, BJ Ferguson3, PM Gressshoff3, S Filleur4, U Mathesius5, R Munns1,6, A Rasmussen7, MH Ryan1, P Ryan6, S Schmidt8, M Watt5
1School of Plant Biology, University of Western Australia; 2Department of Parks and Wildlife, Western Australia; 3Centre for Integrative Legume Research, University of Queensland, 4CNRS, Gif sur Yvette, France; 5Research School of Biology, Australian National University; 6CSIRO Agriculture, Canberra; 7Centre for Plant Integrative Biology, University of Nottingham, UK; 8School of Agriculture and Food Science, University of Queensland
With acknowledgements to authors of the original edition Chapter 3, sections 3, 4 and 5 respectively: BJ Atwell, JWG Cairney and KB Walsh
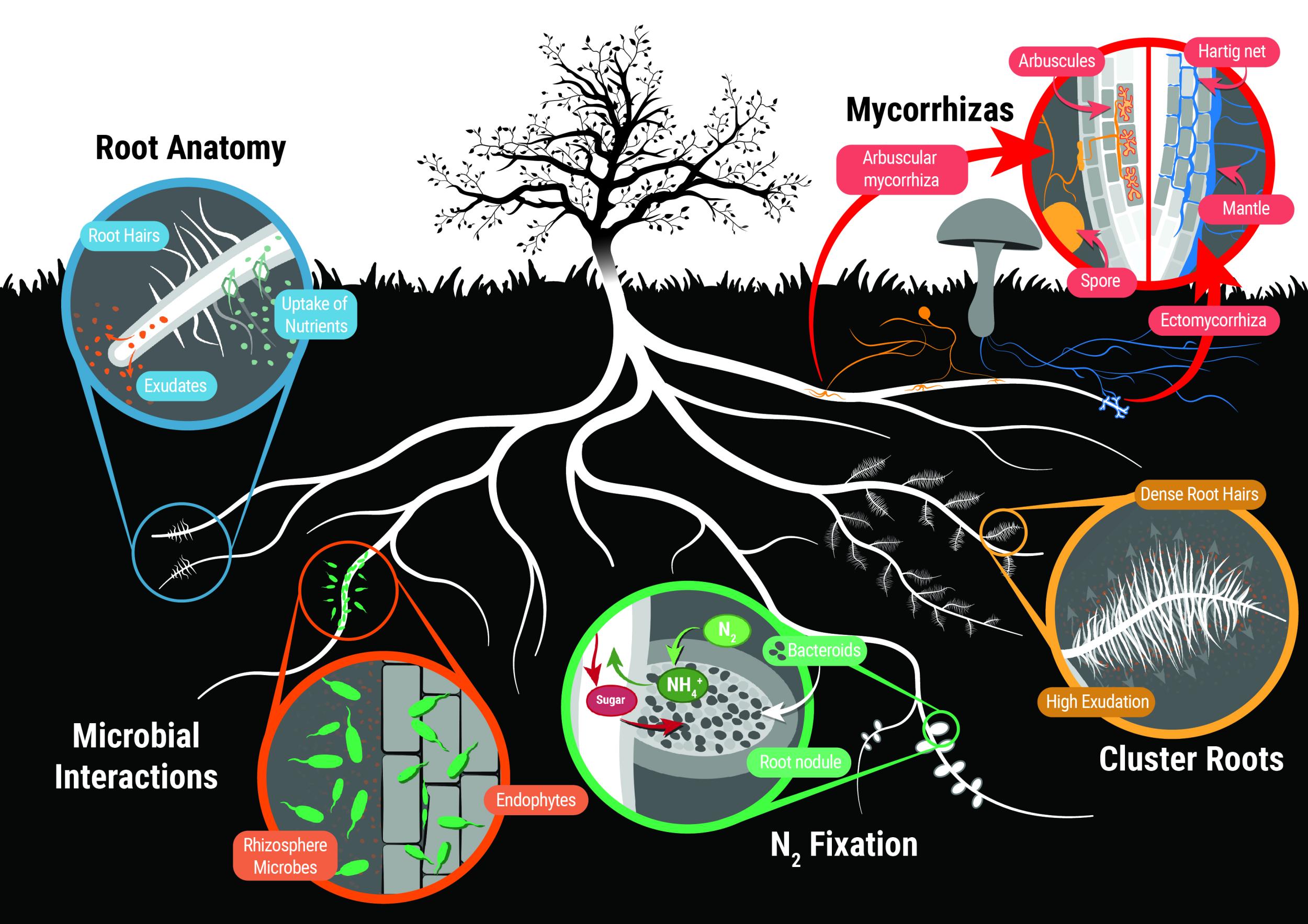
Plants require at least 14 essential minerals for growth, along with water and carbohydrates. The processes by which plants convert CO2 to carbohydrate are described in Chapters 1 and 2 of this text book, and Chapter 3 explains how plants take up water from the soil and transport it to leaves. Chapter 4 describes the fundamental processes by which plants acquire minerals from the soil, with N and P as main examples. In most situations, roots do not take up minerals directly from the soil, but work in association with soil microbes that make the minerals more available to plants.
This chapter first explains the concept of plant nutrition, then describes the various root structures and symbioses with microorganisms that allow plants to take up essential nutrients. These adaptations include specialised root architecture, cluster roots, rhizosphere organisms, mycorrhizas, and symbiotic nitrogen fixation. Finally the principles of membrane transport which require ATP hydrolysis or specialised membrane transporters are described with a focus on uptake of nitrate, different forms of organic N, and phosphate.
Nutrient application to soils via fertilizers, and the ecophysiology of nutrient relations are covered in Chapter 16.
Amanda Rasmussen1 and Susanne Schmidt,2
1Centre for Plant Integrative Biology, University of Nottingham, UK; 2School of Agriculture and Food Science, University of Queensland
Plants require at least 14 essential elements called ‘mineral nutrients’ to sustain life function and complete their life cycle, in addition to carbon (in the form of CO2), oxygen, and hydrogen (in the form of water). Some plants have specific requirements for additional elements. The acquisition via the roots and use of these elements are the topic of plant nutrition.
Nutrients are taken up by roots via active or passive transport across membranes, and travel from the bulk soil to the roots via diffusion or mass flow. However, in order to access all the available nutrients, plants have evolved dynamic and plastic root systems that explore the soil for maximum nutrient uptake. In monocots, lateral roots grow into the volume of soil between seminal roots, as shown by in situ CT imaging (Figure 4.1).By responding to signals and gradients in the soil, the root system can maximise growth in local nutrient patches while minimising growth in areas of deficiency. This is extremely important for plant survival particularly in deficient or marginal soils. Efficient root growth is also an important factor in maximising yield with lower fertiliser applications because ‘wasted’ root growth costs energy that could otherwise be invested in the crop of interest (whether seeds, leaves, stems or tubers). For this reason understanding the root environmental responses and breeding crops with efficient root systems for the conditions of interest are currently highly active areas of agronomic research.
This section covers the different nutrients required for plant growth, and the different root architectures and structures which help the plants maintain sufficient nutrient uptake to support the above ground biomass.
Although the absolute quantities of nutrients required vary between plant species, genotypes and growth environments, essential nutrients are categorised into so-called macronutrients (N, K, Ca, P, Mg, S) that plants require in larger quantities, and micronutrients (Fe, Cl, Mn, B, Zn, Cu, Mo, Ni) that are needed in small amounts (Figure 4.2). Additional beneficial elements include Si (e.g. for grasses) and Na (for many sea-shore plants).
Macronutrients form the structural components of proteins, cell walls, membranes, nucleotides and chlorophyll, and have roles in energy and water maintenance. The macronutrient potassium has a special function in regulating the osmotic potential of plant cells. Under saline or dry conditions, Cl (and for some plants Na) is important in plant water relations.Micronutrients mainly provide functional groups in enzymes (BOX 1 shows how Ni forms the active site in urease, as an example).
BOX 1 – Nickel (Ni) at the centre of UreaseIn 1926 James B. Sumner from Cornell University studied the structure of Urease from Jack Bean plants and demonstrated that the enzyme is also a protein. This work led to the recognition that most enzymes are in fact proteins and in 1946 Sumner was awarded the Nobel Prize in Chemistry. Urease is an enzyme that breaks down urea to ammonia and carbon dioxide in plants, bacteria and fungi and contains a nickel active site. (NH2)2CO + H2O -> CO2 + 2NH3 For 3D structure see http://www.proteopedia.org/wiki/index.php/Urease Further reading: Follmer 2008; Carter et al. 2009. |
In terrestrial ecosystems and in agriculture, the availability of nitrogen (N) and phosphorus (P) are often limiting and so affect plant growth and productivity most strongly. However, other elements can also be limiting. Plants showing nutrient deficiencies will exhibit symptoms such as stunted growth, leaf or shoot tip chlorosis, and defoliation, and will die if supplements are not provided. Fertilisers are applied to supply essential elements in agriculture to maximise plant growth and enhance yields. Along with the discovery of ‘dwarfing genes’ and development of short stature crop varieties, it was the use of synthetic nitrogen fertilisers that played a significant role in the Green Revolution of the 1930s-1960s.
The acquisition of mineral nutrients starts with their movement from the surrounding soil to root surfaces. The movement of nutrients from bulk soil towards plant roots occurs via diffusion or mass flow (Figure 4.3). Root interception occurs as the root comes in contact with, and displaces the soil through which it is growing. Nutrient availability in soils and the physical and chemical factors influencing their movement from soils to the root surface is comprehensively described in a review by Marschner and Rengel (2012). Nutrients are taken up into roots by active or passive transport across cell membranes, which is described later in this chapter (Section 4.5).
Diffusion occurs along a concentration gradient, over relatively short distances (in the order of 1 cm). As roots take up nutrients and ions from the soil a depletion zone can be established allowing diffusion to occur into the depletion zone. The rate of diffusion depends on how fast the roots are taking up the nutrient, how much of the nutrient is present in the soil (this determines the steepness of the concentration gradient that forms) and also on the mobility of the ions by diffusion. Soluble ions would take about a day to diffuse 1 cm; ions bound to the soil matrix would take longer. For examples, Marscher and Rengel (2012) show that nitrate by diffusion in a ‘typical’ soil travels 3 mm in a day, potassium about 1 mm in a day, and phosphate moves only about 0.1 mm in a day. This illustrates the importance of root hairs in intercepting and accessing phosphate.
Mass flow is driven by the uptake of water caused by the transpiration rate of the plants and can occur over long distances. Many soluble nutrients such as nitrate are dissolved in the soil water and as the plant pulls the water from the soil, the nutrients move too. Some nutrients move by mass flow faster than their uptake rate so they build up on the surface of the root during daylight hours (Marschner and Rengel, 2012). The rate of movement by mass flow of solution depends basically on the rate of transpiration of the plant, so there is little movement at night. It is also influenced by soil water content and soil texture (see Chapter 3, Section 3.4).
Nutrients are unevenly distributed in the soil, generally being concentrated in the topsoil due to decomposition of leaf litter, but also dispersed elsewhere in pockets. Uneven surface enrichment arises from diverse sources such as dead fauna, urine patches from grazing animals, and localised application of fertiliser. Phosphorus and all cations are relatively immobile as they bind to the soil while nitrate and other anions (except phosphorus) are soluble and can readily be leached to deeper soil layers.
Because the soil is so heterogeneous, plants have developed adaptable (plastic) root systems so that the roots proliferate close to the nutrients for uptake.
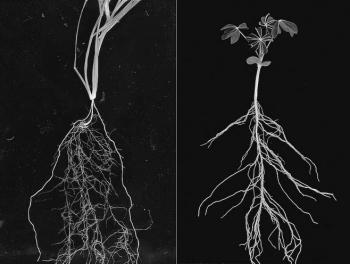
Figure 4.4 Root systems of young (left) wheat and (right) lupin plants. Wheat, a monocot, has a dual root system. Seminal roots emerge from the seed and nodal roots (thicker roots on the outside of the picture) emerge from the crown, a group of closely packed nodes from which tillers emerge. Lupin, a dicot, has a tap root from which lateral roots emerge and which thickens with time as continued cambial activity leads to secondary growth.
The root system architecture is the arrangement of different roots in solid space. Just like a building has walls, roof, and floors, plant root systems also contain different structures including root types (primary, lateral, adventitious), root hairs, and specialized features such as nodules and cluster roots (see case study). In contrast to a fixed structure like a building, the root system is dynamic with new structures forming as needed to explore the soil and old structures breaking down when their use has expired. This four dimensional architecture within soil can now be visualized using technology such as X-ray microscale computed tomography (microCT) and magnetic resonance imaging (MRI).
In order to understand root architecture it is important to understand the different structures that make up the root system. This section will focus on the different root types while cluster roots are explained in the case study that follows. Root hairs are described in the section on the rhizosphere (Section 4.2) and the formation of N2-fixing nodules in Section 4.4.
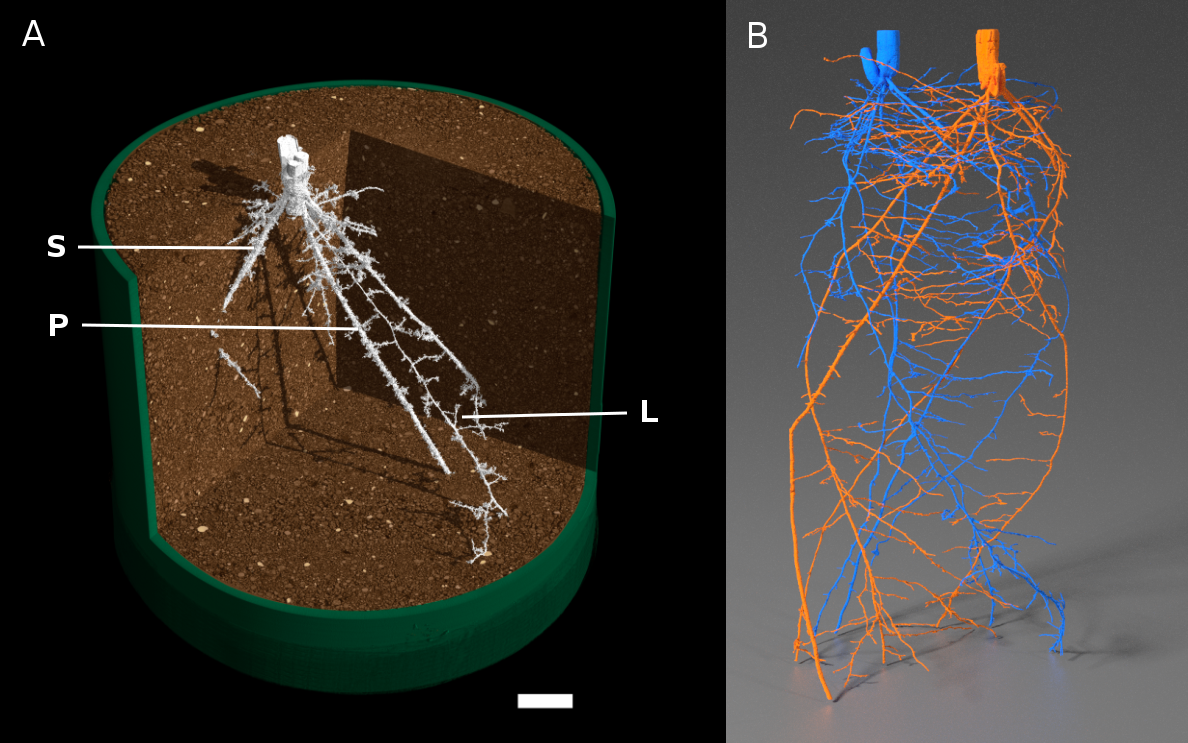
Figure 4.5 MicroCT images of crop root systems. A, Single wheat plant with primary root (P), lateral roots (L) and seminal roots (S). Roots are false-coloured in white, and soil is false-coloured in brown. Scale bar = 1 cm. (Reproduced from Atkinson et al. (2014) Plant Physiol 166: 538-550. doi:10.1104/pp.114.245423. Copyright American Society of Plant Biologists). B) Two wheat plants grown in the same pot. Roots are false-coloured in blue or orange; each colour shows a single root system. (Image courtesy S. Mairhofer)
Lateral roots: roots that form from other roots. The lateral roots that form from the primary root are first order lateral roots; the lateral roots that form from the first order laterals are second order laterals and so on. This class of root is post-embryonic.
Seminal roots: form adjacent to the radicle and dominate the early root growth in monocots. This root type is embryonic.
Adventitious roots: any root that forms from anything other than another root. This includes roots that form on the base of stem cuttings, from leaf explants, from stems in flooded plants and also from nodes of cereal crops (often called crown roots). These root types are very diverse (Steffens and Rasmussen, 2015) so can include both embryonic and post-embryonic roots.
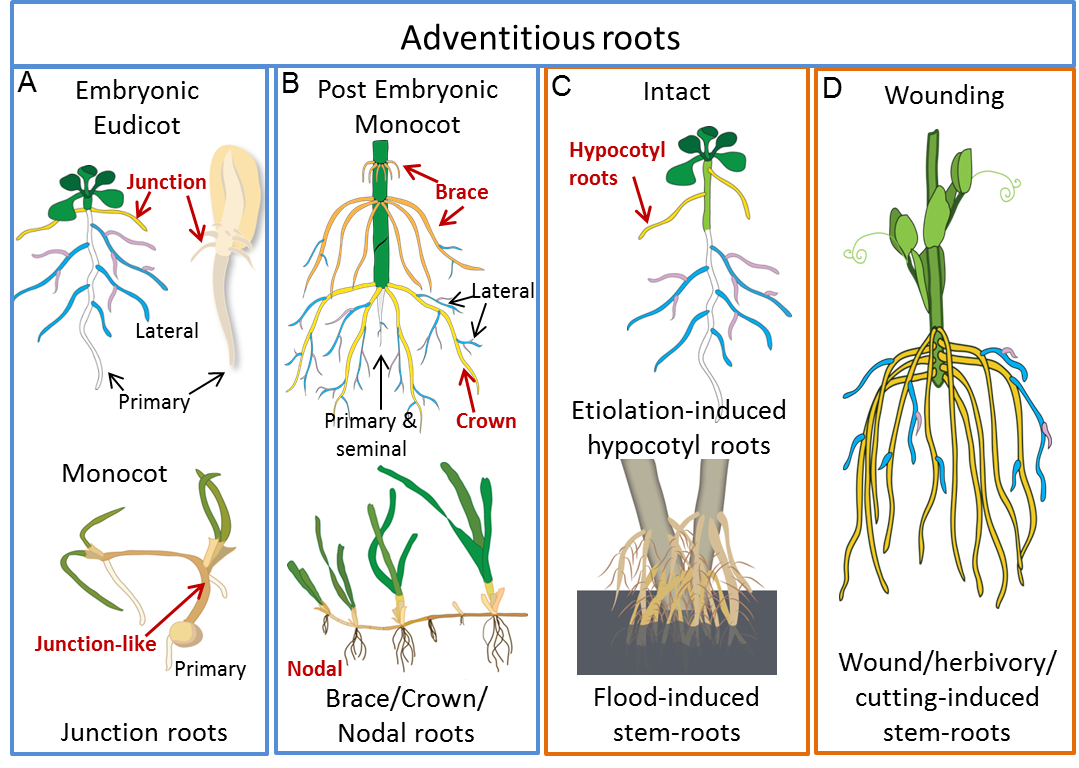
Figure 4.7 Examples of adventitious root types. This figure highlights a few examples of the diversity of adventitious roots; A and B show types of adventitious roots that form during normal development while C and D are examples of stress-induced adventitious roots. A, Those potentially established in the embryo. B, The dominant root system of monocots including maize (top image) crown roots (yellow) and brace roots (orange) and nodal roots on other grasses (lower image) and on eudicots such as strawberry. C, Low or no light (e.g. Arabidopsis used as a model for adventitious root regulation) or flooding (lower image) can induce adventitious roots from either nodal or non-nodal stem positions. D, Wounding such as taking a cutting induces de novo adventitious root development. Primary and seminal roots are depicted in white, first order lateral roots in blue and second order laterals in pink. (Based on Steffens and Rasmussen (2016) Plant Physiol. 170: 603-617. doi:10.1104/pp.15.01360. Copyright American Society of Plant Biologists)
Root hairs: single-cell, hair-like extrusions from the epidermis which increase root surface area for nutrient uptake (Jones and Dolan, 2012) and are important for nodulation (Section 4.4).
The combination of different root types present in the root system differs across species. In particular the root systems of cereal crops (monocots) differ dramatically to the root systems of tree crops (eudicots) (Figure 4.8).
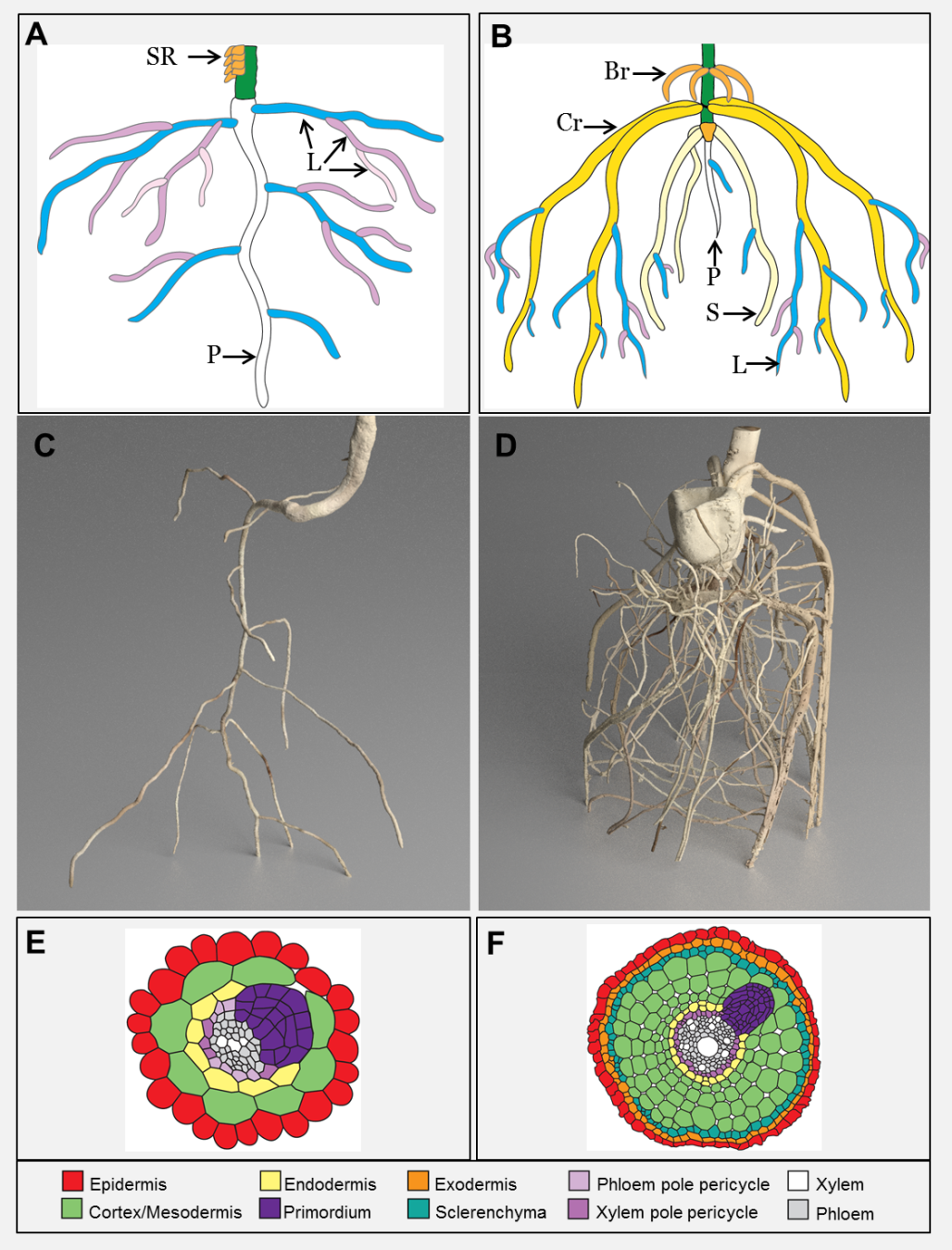
Figure 4.8 Eudicot and monocot root systems. A ,C and E represent eudicot roots while B, D and F represent monocot roots. Schematic showing the dominant root types of tomato (A) and maize (B). SR = stem roots, P = primary root, L = lateral root, Br = brace root, Cr = crown root, S = seminal root (original diagram courtesy A. Rasmussen). MicroCT images of tomato (C) and maize (D) (images courtesy J. Johnson and S. Mairhofer). Cross section schematics of the eudicot Arabidopsis (E) and rice (F) showing where new roots initiate. (Reproduced from Atkinson et al. (2014) Plant Physiol 166: 538-550. doi:10.1104/pp.114.245423. Copyright American Society of Plant Biologists)
Eudicots typically develop a primary (tap) root from a single radicle that emerges from a seed. This primary root, plus the first order lateral roots which emerge from it, provide a framework on which higher-order lateral roots are formed. Such a framework strengthens due to secondary thickening when division of the cambium gives rise to more cell layers, leading to massive roots that are often seen radiating from the base of a tree trunk (Figure 4.9).
Monocots such as grasses and cereal crops do not have a cambium for secondary thickening and develop a fibrous root system. This root system begins with the radicle which grows into the primary root. Adjacent to the radicle, several seminal roots also emerge and combined with the primary root these roots dominate the young root systems of monocots. Next nodal adventitious roots (often called crown roots) emerge from lower stem nodes and these thicker roots gradually dominate the root system. Finally in some monocots, such as maize (corn), nodal adventitious roots emerge above the soil level (brace roots) to provide additional structural support. Stems of monocots are typically anchored by the nodal roots, which are more numerous than seminal roots (Hochholdinger et al. 2004; Hochholdinger 2009).
Despite these structural differences between monocot and eudicot root systems, they can all vary the soil volume which they explore depending on water and nutrient availability. In this way the root distribution in the soil can vary both vertically and locally depending on available resources.
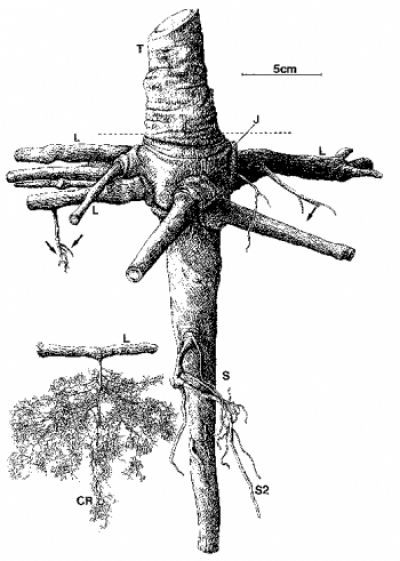
Figure 4.9 Dimorphic root system of a six-year-old Banksia prionotes tree growing in Western Australia in a deep sand. The trunk (T) is connected through a swollen junction (J) to the root system which comprises a dominant sinker root (S) with smaller sinkers (S2). A system of lateral roots (L) emerge horizontally from the junction, some bearing smaller sinker roots (arrows). Other laterals give rise to cluster roots (CR). (W.D. Jeschke and J.S. Pate, J Exp Bot 46: 907-915, 1995)
The amount of roots present in a volume of soil varies both vertically and locally depending on resource availability and physical restrictions. This is often measured as the total length of all roots present per unit volume of soil (root length density, L, expressed in km m–3).
Vertically, the root length density is often large in surface layers of the soil and typically decreases with increasing depth. Commonly, hundreds of kilometres of root per cubic metre of soil are observed near the soil surface.
Figure 4.10 shows root length density, L, as a function of depth in a wheat crop in early spring, and under a jarrah forest, also in spring. Both have a dense population of roots near the surface but wheat roots barely penetrate below 1 m, whereas jarrah roots penetrate to well below the 2.5 m shown here, often to 20 m. Dense root proliferation near the soil surface probably reflects an adaptation of plants to acquire phosphorus, potassium and other cations such as the micronutrients zinc and copper. These nutrients do not move readily in soil as they are bound to the soil surfaces, hence roots branch prolifically to ensure close proximity (a few millimetres) between adsorbing surfaces and these soil-immobile ions. Roots of jarrah are also concentrated near the soil surface (Figure 4.10) to access phosphate and nutrients released by litter decomposition, but some roots penetrate very deeply to tap subsoil moisture.
Nutrients are distributed unevenly in the soil. Root systems respond to enriched zones of nutrients by high levels of branching. Figure 4.11 shows an example of such a proliferation; the dense roots in the centre of the figure are a response by the row of wheat plants to application of a large pellet of nitrogen fertiliser (see arrow).
Such proliferations around bands of fertilizer ensure plants maximise nutrient uptake with the minimum cost to plant development. This efficiency fits within optimal partitioning theory which states that plants respond to environmental variation by partitioning biomass among the plant organs to optimize the acquisition of nutrients, light, water and carbon to maximize plant growth (Reich, 2002). This means that in low nutrient conditions the plants will put more energy into growing roots and less into shoot growth (Reich, 2002). Likewise when light is limiting, plants will invest more energy in leaf area and less in root development (Weaver and Himmel, 1929; Reich, 2002). Maintaining the balance between root and shoot is important as the roots must be extensive enough to supply nutrients and water in proportion to the demand and hydraulic pull from the leaves and vice versa the leaves must produce enough sugars to continue the growth of the root system (Weaver and Himmel, 1929). Consistent with this, Butler et al. (2010) found in Sitka spruce forest, the root absorbing area was correlated with the tree stem diameter and to the transpiring leaf area index. This highlights the link in hydraulics between leaf and root areas.
Young roots absorb nutrients more rapidly than old roots. New roots supply annual plants with abundant sites for nutrient uptake, especially during establishment. A feature of the roots of perennials is that they have a large turnover of the fine, high-order lateral roots that emerge from the secondarily thickened framework each year. This turnover draws heavily on photoassimilate, equivalent to half the CO2 fixed in annuals and up to 90% of the standing biomass of temperate forests. Production of fine (and often ephemeral) roots ensures uptake of nutrients over many years.
Because many soils are deficient in key nutrients, plants have developed a special relationship with certain fungi called mycorrhizae (Section 4.4). In this symbiosis the fungi obtain fixed carbon from the host plant, and in turn supply the host with poorly mobile nutrients, especially phosphorus. This is achieved by proliferating their hyphae to provide a much greater surface area for nutrient uptake than could be provided by roots alone. Another adaptation, common in the Proteaceae, and also occurring in some species of lupin, is proteoid roots, clusters of tiny rootlets that greatly enlarge the available surface area for ion uptake and which are inducible by low levels of phosphorus (see Case study 4.1).
M. Watt
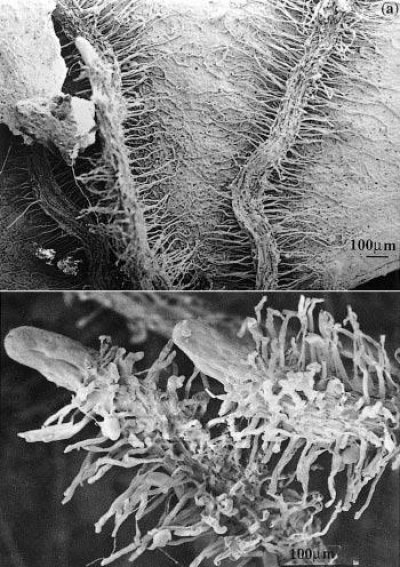
Figure 1 Cluster roots in Banksia serrata growing on Hawkesbury Sandstone hillslopes in the Sydney region. a, Roots that have grown across a dead eucalypt leaf extract nutrients remaining in the decaying leaf. b, Clusters of fine rootlets at the tips of roots increase the surface area for nutrient extraction from surrounding soil. Scale bar = 100 µm. (Scanning electron micrograph courtesy S. Gould)
Australian soils generally contain low concentrations of plant-available phosphate, much of it bound with iron–aluminium silicates into insoluble forms or concentrated in the remains of decaying plant matter. Because very little of this phosphate is soluble, most roots extract it only slowly. Plants with cluster roots gain access to fixed and organic phosphate through an increase in surface area and release of phosphate-solubilising exudates. Hence plants with cluster roots grow faster on phosphate-fixing soils than species without clusters.
Cluster roots have a distinct morphology. Intense proliferation of closely spaced, lateral ‘rootlets’ occurs along part of a root axis to form the visually striking structures. Root hairs develop along each rootlet and result in a further increase in surface area compared to regions where cluster roots have not developed.
In the Proteaceae, clusters generally form on basal laterals so that they are abundant near the soil surface where most nutrients are found. For example, Banksia serrata produces a persistent, dense root mat capable of intercepting nutrients from leaf litter and binding the protecting underlying soil from erosion (Figure 1a). New clusters differentiate on the surface of this mat after fires and are well placed to capture nutrients. In contrast, Banksia prionotes forms ephemeral clusters which export large amounts of nutrients during winter. Lupinus albus has more random clusters which appear on up to 50% of roots (Figure 2).
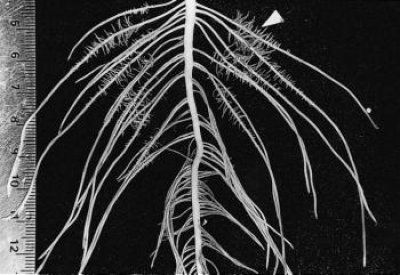
Figure 2 Basal roots of a two-week-old Lupinus albus plant grown in nutrient culture with 1 µM phosphate. Proteoid roots have emerged along the primary lateral roots (arrowhead) and the oldest proteoid rootlets have reached a determinate length of 5 mm. As rootlets approach their final length, they exude citrate for 2-3 d. (mm scale on left side) (Photograph courtesy M. Watt)
Rootlets not only represent an increase in surface area but also exude protons and organic acids, solubilising phosphate and making it available for uptake (Watt and Evans 1999a). Exudates from cluster roots represent up to 10–23% of the total weight of an L. albus plant, suggesting that they constitute a major sink for photoassimilates. However, not all this additional carbon comes from photosynthesis because approximately 30% of the carbon demand of clusters is met by dark CO2 fixation via phosphoenolpyruvate carboxylase. Because cluster roots form on roots of L. albus even when phosphate supply is adequate, growth of L. albus in soils with low phosphate availability is not restricted by an additional carbon ‘drain’ to roots. On the other hand, the great many species which produce cluster roots in response to environmental cues like phosphate deficiency might experience a carbon penalty to support these roots.
Cluster roots on L. albus are efficient with respect to carbon consumption by generating citrate on cue. Most of the citrate exuded by clusters is released during a two to three day period when the cluster is young (Watt et al. 1999b). A large root surface area in clusters works in concert with this burst of exudation to solubilise phosphate before it is re-fixed to clay surfaces or diffuses away.
Cluster roots can mine a pocket of phosphate-rich soil which would otherwise not yield its nutrients. They are an elegant adaptation of root structure and biochemistry to nutrient-poor soils.
Dinkelaker B, Hengeler C, Marschner H (1995) Distribution and function of proteoid roots and other root clusters. Bot Acta 108: 183–200
Lamont BB (1993) Why are hairy root clusters so abundant in the most nutrient impoverished soils of Australia? Plant Soil 156: 269–272
Purnell HM (1960) Studies of the family Proteaceae 1. Anatomy and morphology of the roots of some Victorian species. Aust J Bot 8: 38–50
Watt M, Evans JR (1999a) Proteoid Roots. Physiology and Development. Plant Physiol 121: 317–323
Watt M, Evans JR (1999b) Linking development and determinacy with organic acid efflux from proteoid roots of Lupinus albus L. grown with low phosphorus and ambient or elevated atmospheric CO2 concentration. Plant Physiol 120: 705–716
Ulrike Mathesius, Research School of Biology, Australian National University
As a general rule, the surface area of a root system exceeds the leaf canopy it supports. Even disregarding root hairs, the interface between roots of a three-week-old lupin plant and soil is about 100 cm2 while a four-month-old rye plant under good conditions has more than 200 m2 of root surface (Dittmer 1937). Trees’ root systems are difficult to quantify but kilometres of new roots each year generate hundreds of square metres of root surface. Such a root–soil interface arises through the simultaneous activity of up to half a million root meristems in a mature tree.
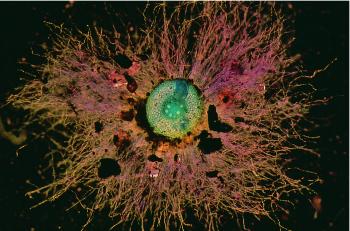
Figure 4.12 Transverse view of a young, soil-grown wheat root, sectioned by hand and stained with Toluidine Blue. Most soil in the rhizosheath was washed away during preparation, revealing many long root hairs extending from the main axis (diameter 0.6 mm). Root hairs allow this root to explore 21 times more soil volume than would be possible without hairs. A lateral root can be seen extending from the pericycle which surrounds the stele. (Photograph courtesy M. Watt)
Many roots form fine extensions to epidermal cells called root hairs, amplifying the effective surface area of the soil–root interface many times. Dittmer (1937) estimated that the surface area of root hairs in rye plants was more than that of the root axes on which they grew; similar observations have been made for trees. The aggregate length of root hairs in the rye plants studied by Dittmer increased 18 times faster than that of the main axes. Thus, up to 21 times more soil is explored when root hairs are present (Figure 4.12).
Root hairs are particularly important in taking up mineral nutrients that are not readily soluble and therefore not mobile in the soil solution, like phosphate. Measurements of the phosphate concentration in soil at different distances from roots show that soil phosphate is depleted only in the zone close to roots, the 1 mm zone, the typical length of root hairs (Figure 4.13).
Anchorage and extraction of inorganic soil resources both call for a large area of contact between roots and soil. However, this vast interface is much more than a neutral interface; events within it allow resources to be extracted from the most unyielding soils. Intense chemical and biological activity in a narrow sleeve surrounding roots, particularly young axes, give rise to a rhizosphere, the volume of soil influenced by the root, a concept first introduced in 1904 by Lorenz Hiltner (Hartmann et al. 2008). The rhizosphere has been estimated to contain up to 1011 microbial cells per gram of soil, and harbour up to or above 30,000 different microbial species, undoubtedly the moxt complex ecosystem on earth (Berendsen et al. 2012). The rhizosphere concept has been extended to include symbiotic mycorrhizal fungi associate with the root (See Section 4.3 on mycorrhiza), and this has been named the ‘mycorrhizosphere’, as most land plants are colonised by mycorrhizal fungi most of the time.
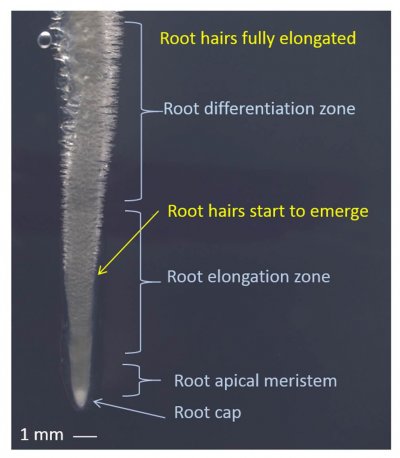
Figure 4.13 Young root tip with elongating root hairs. Root tip of Medicago truncatula showing the approximate zones of root elongation and differentiation relative to the tip. Root hairs are protuberances of epidermal cells that first emerge approximately 4 mm behind the root tip and elongate over the span of about two days until they are fully elongated in the differentiation zone of the root. (Photograph courtesy U. Mathesius)
The rhizosphere is the narrow zone of soil surrounding plant roots that is characterised by root exudation and an abundance of micro-oganisms which can be beneficial or harmful to plants, or have no effect on root growth and function. These microbes are saprophytic, pathogenic or symbiotic bacteria and fungi, including rhizobia forming nodules and arbuscular mycorrhizal fungi (Figure 4.14).
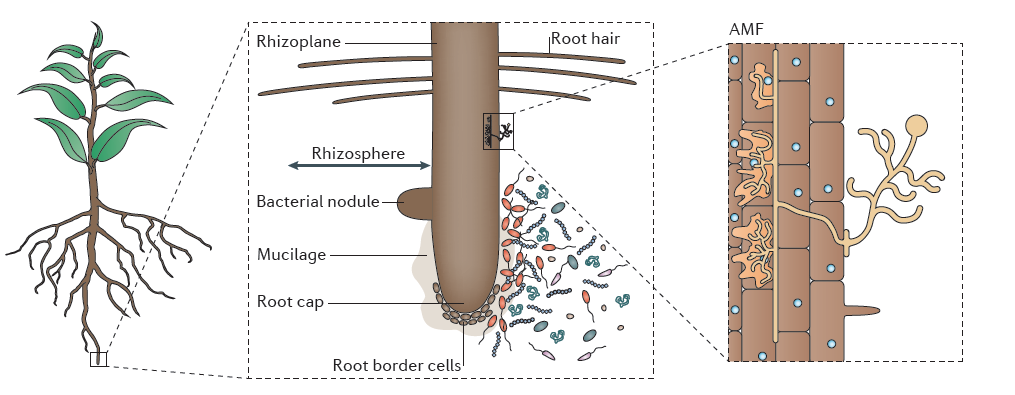
Figure 4.14 The rhizosphere is the narrow zone of soil surrounding plant roots that is characterised by root exudation and an abundance of saprophytic, pathogenic and symbiotic bacteria and fungi. These include rhizobia that form nodules, and arbuscular mycorrhizal fungi (AMF). The rhizoplane describes the root surface in contact with the soil. Root cap and root border cells near the root tip provide lubrication as the root expands into the soil. (Reproduced by permission from Macmillan Publishers Ltd from L. Philippot et al. Nature Rev Microbiol 11: 789-799, 2013)
Many root phenomena suggest specific roles for the rhizosphere. For example, roots have long been thought to find a relatively frictionless path through soils because of exudation of organic substances and cell sloughing, but the chemical and physical processes that underpin this phenomenon are still quite unclear (McKenzie et al. 2012). Production of new roots around local zones of enrichment (Section 4.1) is made far more effective through rhizosphere activity associated with these young roots. Phosphate availability is particularly likely to be improved by the presence of a rhizosphere. Potential mechanisms will be discussed below.
Enhancement of root growth under conditions which favour high root:shoot ratios and the attendant rhizosphere surrounding those roots (rhizosheath) require a substantial input of organic carbon from shoots. Some is used in structural roles, while roots and microbes also require large amounts of carbon to sustain respiration. Even in plants growing in nutrient-adequate, moist soils, 30–60% of net photosynthate finds its way to roots (Marschner 1995). Carbon allocation to roots can be even greater in poor soils or during drought. The rhizosphere accounts for a large amount of the root carbon consumption (Jones et al. 2009). Barber and Martin (1976) showed that 7–13% of net photosynthate was released by wheat roots over three weeks under sterile conditions while 18–25% was released when roots were not sterile. This difference might be considered carbon released because of microbially-induced demand in the rhizosphere, and therefore made unavailable for plant growth.
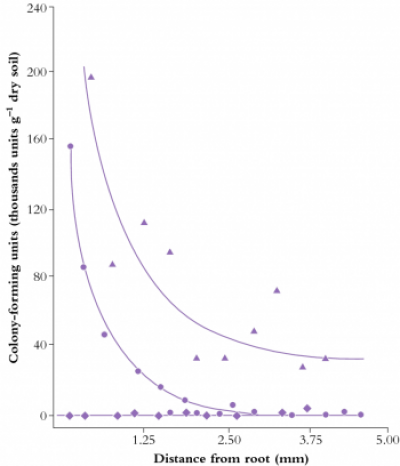
Figure 4.15 Concentration of Enterobacter cloacae (RP8) around wheat roots when the bacterium was introduced by inoculating seeds (circles) or soil (triangles). Uninoculated controls are shown as diamonds. Approximately 3 mm of the soil around roots supports an elevated bacterial population (A.F. Dijkstra et al. Soil Biol Biochem 19: 351-352, 1987)
Rhizosphere chemistry and physics differ from the adjacent soil matrix and root tissues. Gradients in solutes, water and gases combine with microbial activity to produce a unique compartment through which roots perceive bulk soil. This zone of influence typically extends not more than 3 mm from the root axis (Figure 4.15), partly due to the low diffusion coefficients of most solutes that move through the rhizosphere (10–12 to 10–15 m2 s–1 for ions such as orthophosphates). Even a relatively mobile ion such as nitrate, with a diffusion coefficient (D) of around 10–9 m2 s–1 in soil solution, diffuses through about 1 cm of soil in a day. Because the time required (t) for diffusion of ions is a function of the square of distance traversed (l), where t = l2/D a nitrate ion would take four days to travel 2 cm, nine days to travel 3 cm and so on. Similarly, organic carbon diffuses away from roots only slowly, sustaining a microbial population as it is consumed in the rhizosphere.
Roots advancing through soil perceive a wide range of chemical and biological environments: a rhizosphere simultaneously fulfils buffering, extraction and defence roles allowing roots to exploit soils. A rhizosphere is thus a dynamic space, responding to biological and environmental conditions and often improving acquisition of soil resources. New roots develop an active rhizosphere which matures rapidly as the root axis differentiates.
Photoassimilate diffuses from roots into the rhizosphere where it is either respired by microorganisms, volatilized, or deposited as organic carbon (‘rhizodeposition’). Some of this photoassimilate loss is in the form of soluble metabolites, but polymers and cells sloughed off the root cap also provide carbon substrates. Grasses undergo cortical cell death as a normal developmental process, providing further carbon substrates to support a rhizosphere microflora. Nitrogen and some other inorganic nutrients which are co-released with plant carbon are often reabsorbed by roots. Extraction of minerals from bulk soil also relies strongly on rhizosphere processes, especially near the root apices. Compounds exuded from roots interact with soil components in direct chemical reactions (e.g. adsorption reactions), through microbially mediated events (e.g. immobilisation reactions) and volatilisation. In addition, complex polysaccharides and glycoproteins of microbial and root origin give rise to a gelatinous mucilage which associates with soil particles to form a rhizosheath.
The rhizosheath is known as the soil that adheres to roots when they are removed from the pot or field Figure 4.16). The amount of soil can vary depending on how gently or roughly the roots are removed. For wheat, at least, the size of the rhizosheath correlates with root hair length. Mutants without root hairs have no rhizosheath. The distinction between the terms rhizosheath and rhizosphere are that the first term refers to the soil that physically adheres, and the second term the volume of soil influenced by the root. Mutants without root hairs would still have a rhizosphere of sorts since the root would still chemically influence its surrounding soil.
Rhizosheaths have physical and chemical implications for root function. Hydraulic continuity between soil and roots is, for example, thought to be enhanced by the hydrated mucigel, which facilitates water uptake by roots in dry soils. Negatively charged groups on side-chains of mucilagenous polysaccharides attract cations like Ca2+, providing exchange sites from which roots might absorb nutrients. The mucigel between the sloughed root cap and root border cells also acts as a lubricant for reducing penetration resistance of the expanding root tip in soil (McKenzie et al. 2012). For example, root elongation through hard soil is greatly reduced if the root cap cells are removed. Once the soil and the mucigel dry up, this lubrication effect is significantly reduced.
Such a diversity of chemical reactions in the rhizosphere is largely an outcome of the array of root-derived exudates. For example, phenolic compounds can be released by root cells in large amounts (Marschner 1995), both as a result of degradation of cell walls and from intracellular compartments. Flavonoids are a group of phenolics that can be specifically exuded into the rhizosphere as signal molecules to attract rhizobia (See section 4.4 on nitrogen fixation). Release of organic acids (principally citric, fumaric and malic acids) solubilises phosphate from surfaces to which they are adsorbed in many species, including those of the family Proteaceae. A modest release of organic acids accounting for about 0.1% of the root mass each week is sufficient to enhance phosphate acquisition in a selection of annual legumes (Ohwaki and Hirata 1992). In more extreme cases, up to a quarter of the dry weight of Lupinus albus plants is released from cluster roots, mostly as citrate (see Case study 4.1). Even the fungal hyphae of mycorrhizal eucalypt and pine roots can secrete photoassimilates, in the form of oxalic acid, causing phosphorus to be solubilised from insoluble calcium apatite (Malajczuk and Cromack 1982).
The main families of low molecular weight compounds which react with inorganic ions are phenolics, amino acids and organic acids. Heavy metals such as aluminium, cadmium and lead are complexed by phenolics, affecting the mobility and fate of these ions in contaminated soils. Flavonoids can chelate iron and make iron oxides available to plants. Manganese is complexed by organic acids, as are ferric ions, which also interact chemically with phenolic compounds and amino acids. For example, highly specialised amino acids (phytosiderophores) can complex ferric ions and enhance uptake from soils by rendering iron soluble. Low iron status actually stimulates release of phytosiderophores into the rhizosphere (Marschner 1995). Other metals such as zinc and copper might also be made more available to the plant through the chelating action of phytosiderophores. Chemical processing by chelating agents is dependent on plant perception of nutrient deficiencies, leading to an ordered change in rhizosphere chemistry. A significant demand on photoassimilates is required to sustain chelation of nutrient ions.
Enzymes are also released from roots, particularly phosphatases, which cleave inorganic phosphate from organic sources. The low mobility of orthophosphates means that phosphatases can be an important agent in phosphorus acquisition, especially in heathland soils where the native phosphorus levels are low relative to the phosphorus-rich remnants of decaying plant material.
pH is another important rhizosphere property. Roots can acidify the rhizosphere by up to two pH units compared to the surrounding bulk soil through release of protons, bicarbonate, organic acids and CO2 (Figure 4.17). In contrast, the rhizosphere of roots fed predominantly with nitrate was more alkaline than bulk soil. A distinct rhizospheric pH arises because of the thin layer of intense biological activity close to roots, especially young roots. In addition to proton fluxes, release of CO2 by respiring roots and microbes is likely to cause stronger acidification of the rhizosphere near root apices where respiration is most rapid.
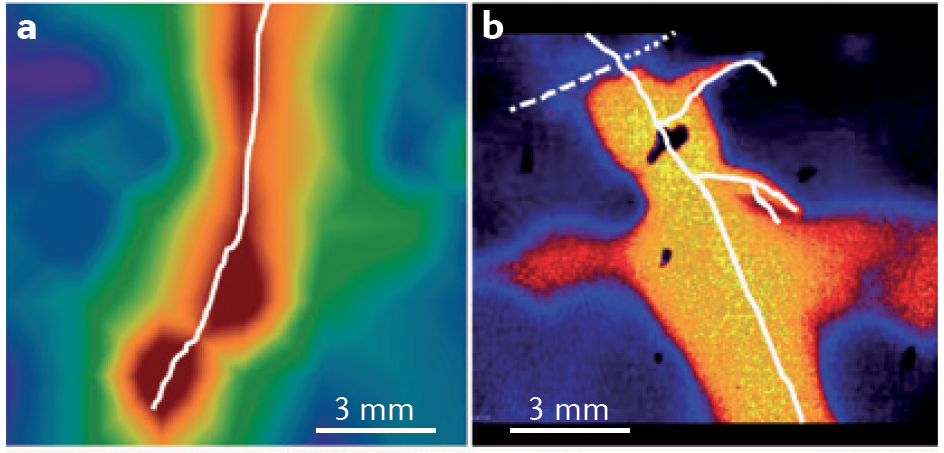
Figure 4.17 Root-induced changes in the rhizosphere. a, Oxygen profiles across a growing root of Juncus effusus (in white). b, pH profiles across growing roots of intercropped durum wheat (dashed white) and chickpea (solid white). (Adapted from L. Philippot et al. Nature Rev Microbiol 11: 789-799, 2013, S. Blossfeld et al. Soil Biol Biochem 43: 1186-1197, 2011, and S. Blossfield et al. Ann Bot 112: 267-276, 2013, with permissions respectively from Macmillan Publishers Ltd, Elsevier, and Oxford University Press)
Rhizosphere acidification affects nutrient acquisition by liberating cations from negative adsorption sites on clay surfaces and solubilising phosphate from phosphate-fixing soils. Furthermore, micronutrients present as hydroxides can be released at low pH, conferring alkalinity tolerance on those species with more acidic rhizospheres. So, the rhizosphere is a space which ensheathes particularly the youngest, most active parts of a root in a chemical milieu of the root’s making. In this way, acquisition of soil resources is strongly controlled by processes within roots. Local variations within soil are buffered by rhizosphere chemistry, enabling roots to exploit heterogeneous soils effectively.
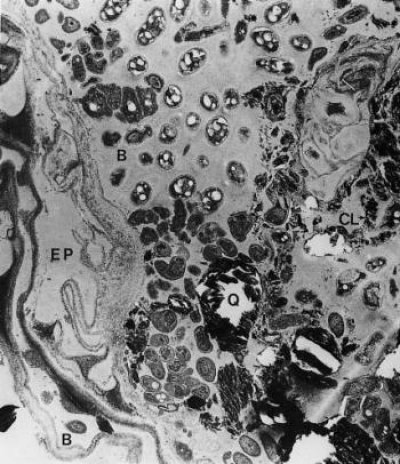
Figure 4.18 Mature rhizosphere from roots of clover (Trifolium subterraneum L.). The outer cortex has been crushed and epidermal cells (EP) have become distorted, leading to leakage of substrates into the rhizosphere. The rhizosphere is rich in microorganisms with bacteria (B) clearly visible. Soil (Q) and clay (CL) particles are held together in the inner rhizosphere by a mucilage of polysaccharides. Sustained losses of carbon required to maintain this microflora are thought to come from exudation and senescence of root cells. (× 10,000) (Courtesy R.C. Foster, A.D. Rovira and T.W. Cock)
Microbial activity, sustained by photoassimilates secreted from roots, contributes substantially to rhizosphere properties. The level of microbial activity is also influenced by availability of nitrogen as a substrate for microbial growth. Soils with high fertility and biological activity have microbial densities 5–50 times greater in the rhizosphere than in bulk soil. The diversity of rhizosphere microflora is spectacular (Figure 4.18) and still incompletely described. An initial hurdle in the identification of rhizosphere microbes was the fact that most of them are unculturable on most known growth media. Metagenomics allows species to be sequenced from soil samples without culturing, largely overcoming this bottleneck. Next Generation sequencing studies of microbes present on root surfaces and in the rhizosphere soil have discovered thousands of different bacterial and fungal species living in close association with plant roots (e.g. Bakker et al. 2013; Bulgarelli et al. 2012; Lundberg et al. 2012). Some of these are very abundant and found in association with many plant species, others are less abundant and highly variable (Figure 4.19).
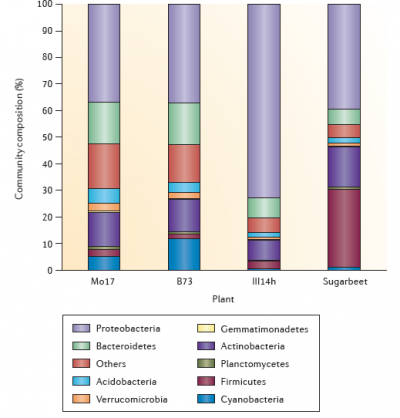
Figure 4.19 The composition of the bacterial community in the rhizosphere. The figure shows examples of the composition of the bacterial community in the rhizosphere of three maize genotypes (Mo17, B73 and III14h) and of sugarbeet. The distribution of the different bacterial phyla is based on data obtained by 454 sequencing (maize) and G3 PhyloChip analyses (sugarbeet). The bacterial community composition was characterized in the rhizosphere of 27 maize genotypes cultivated in five fields located in three states in the USA. Here, three genotypes displaying contrasted rhizosphere microbiota in a given field are depicted for illustration and the sugar beet rhizosphere microbiota presented is from seedlings grown in a disease-conducive soil in The Netherlands. (Reproduced by permission from Macmillan Publishers Ltd from L. Philippot et al. Nature Rev Microbiol 11: 789-799, 2013)
These microbes can almost be viewed as an extension of the plant into the soil. Like the human gut microbiome, the plant rhizosphere microbiome appears to be an essential part of the plant with multiple functions in nutrition and pathogen defense; it is inseparable from the plant and has been dubbed the plants second genome. The rhizosphere community is highly structured and not a random collection of species – it is strongly influenced by plant species and even ecotypes, by the type of the soil, availability of nutrients and the exudation of chemicals from the root (Bakker et al 2013). Plant mutants with altered chemical composition of root exudates have been found to attract significantly altered microbial communities. It will be fascinating to discover to what extent this is an active strategy of the plant to attract the most appropriate rhizosphere microbiome to help the plant survive in a given environment.
Rhizosphere microorganisms are also not uniformly distributed along roots. Apices are almost free of microbes but densities can increase dramatically in subapical zones. Very mature root axes with lateral branches are sparsely populated with microbes. Even within these zones, there are large variations in distribution, with radial epidermal walls of roots secreting exudates which can support huge microbial populations, up to 2 × 1011 microbes cm–3. Composition of microbial communities varies with their distribution along the root as well, likely reflecting different nutrient sources along the root. Fluorescence in situ hybridization (FISH) can be used to visualise different taxa of bacteria on the root surface (rhizoplane; Figure 4.20).
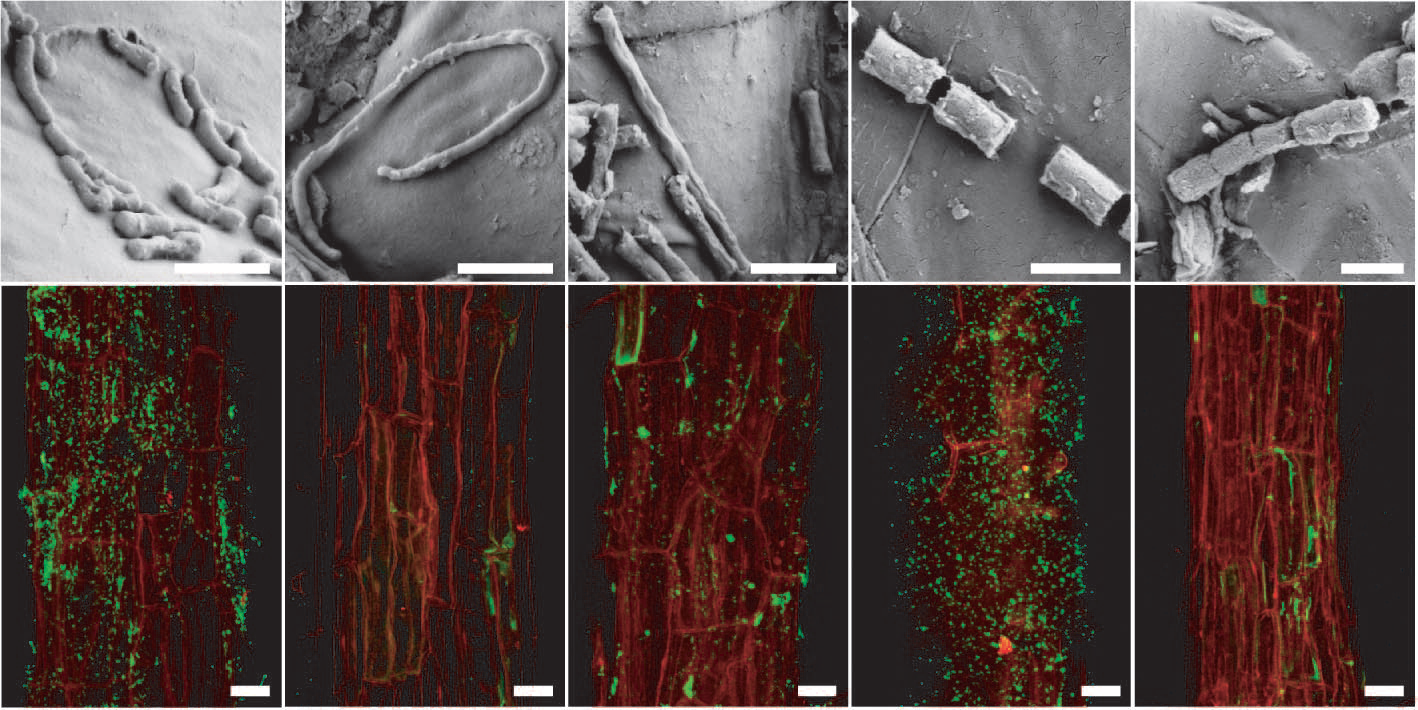
Figure 4.20 Arabidopsis root-inhabiting bacteria are detectable on the rhizoplane. a to e, Scanning electron micrographs of bacteria-like structures. Bars, 1 mm. f to j, Detection of bacteria by fluorescence in situ hydridisation (FISH) using probes against specific bacterial groups (bacteria in green due to AlexaFluor488) on the root surface (red, root autofluorescence) by confocal laser scanning microscopy. f, Most Eubacteria detected with probe EUB338. g, Negative control with reverse complementary probe of EUB338 (NONEUB). h, Betaproteobacteria detected with probe BET42a. i, Bacteroidetes detected with probe CF319a. j, Actinobacteria detected with probe HGC69a. Bars, 20 mm. (Reproduced by permission from Macmillan Publishers Ltd from D. Bulgarelli D et al., Nature 488: 91-95, 2012)
Roots do of course influence adjacent soil throughout their length by setting up gradients of water, gases and ions. For example, in waterlogged soils leakage of O2 from aerenchymatous roots leads to oxidation of metal ions and local build up of aerobic microflora around roots of agricultural plants (Chapter 18). In general, however, the most active microbial populations and rates of chemical transformation in the rhizosphere occur in the subapical zones of the root. In supporting these processes, root-associated microbes metabolise inorganic nitrogen, depositing protein nitrogen in the process of immobilisation. Microbial activity also produces plant growth regulators such as auxin, cytokinins and gibberellins, sometimes in amounts sufficient to influence root morphogenesis. Ethylene can also be produced by rhizospheric fungi, potentially influencing root morphological changes such as lateral root initiation. Some bacteria have been found to promote plant growth by reducing ethylene levels around roots through production of an enzyme degrading an ethylene precursor, 1-aminocyclopropane-1-carboxylate (ACC) deaminase.
Root function and overall plant performance can benefit conspicuously from processes in the rhizosphere. Infection by rhizobia (Section 4.4) and mycorrhizal fungi (Section 4.3) improve the nutritional status of many species. Rhizobial strains have even been used to manipulate rhizosphere biology. A significant proportion of photoassimilate is used to support a rhizosphere, reflecting the high cost of microbial activity and polymer exudation. This pattern is repeated in many species with up to 20% of plant carbon consistently lost by roots, however, this value can vary substantially with the biotic and abiotic conditions. Relative rates of microbial and root respiration are almost impossible to estimate in roots growing in undisturbed soils because of the intimacy of roots and microbes. In addition to consuming large amounts of plant carbon, some microbes can produce phytotoxins, which can impose further restrictions on root function. Some microbes also contribute to nutrient depletion in the rhizosphere, for example by converting usable forms of nitrogen, i.e. nitrate or ammonium, into unusable forms like N2.
Mechanisms describing how a rhizosphere benefits its host are even more elusive because of the diversity of reactions in such a small space. Chelation is identified as a major influence on nutrient acquisition and might also help ameliorate ion toxicities. Physical properties of the rhizosphere are even less well understood, with questions such as root lubrication, root–mucilage shrinkage and interfacial water transport not yet resolved. Physical properties of mucilage do not suggest it is an ideal lubricant. Whether the dynamic properties of a rhizosphere bring constant benefits to a plant or simply passively coexist with growing roots remains a critical question.
One demonstrated benefit of the rhizosphere microbiome is the protection of the plant from diseases. Several mechanisms have been suggested for this effect (Bakker et al. 2013; Berendsen et al. 2012): Disease suppressiveness, the ability of the microbial inhabits of the rhizosphere to suppress the infection of plants by soil-swelling pathogens, has been ascribed to the production of antimicrobial substances by bacteria, to competition between beneficial and pathogenic microbes and to the induction of systemic resistance by beneficial bacteria. An intriguing example is the colonization of plants by pathogens, which can lead to changes in the exudation of organic acids that then attract beneficial bacteria that induce systemic resistance in the plant, reducing pathogen infection. The induction of systemic resistance to pathogens can also be triggered by specific signaling molecules of bacteria, quorum sensing signals, which bacteria used to ‘talk’ to each other to coordinate multicellular-like behaviours of bacterial colonies. Perception of quorum sensing signals from rhizosphere bacteria by plants can increase systemic resistance to pathogens in the shoot, and can also enhance symbiosis with nitrogen-fixing bacteria. Quorum sensing signal perception also triggers the production of so called quorum sensing mimic compounds – signals that interfere with bacterial communication in the rhizosphere (Teplitski et al. 2011). While we still need to identify most of the signals, signal mimics and exudate components in the interaction of roots with their microbiome, it is clear that plants actively create the rhizosphere, and that this is likely to benefit the plant in its environment.
Megan H Ryan1 and Mark C Brundrett1,2
1School of Plant Biology, University of Western Australia; 2Department of Parks and Wildlife, Western Australia.
The roots of around 90% of higher plants form a symbiotic association with mycorrhizal fungi (Figure 4.21). These fungi colonise roots, with the colonised root being termed a “mycorrhiza”. The fungi benefit from the provision of plant carbon. The host plant may benefit in many ways, but the primary benefit is most often the ability to access inorganic nutrients from soil beyond the rhizosphere due to their transport into the root by hyphae of the fungi. Mycorrhizal associations are present in plants in both natural ecosystems and modern agricultural systems; although their occurrence in the latter may be reduced by common management practices, especially the addition of fertiliser.
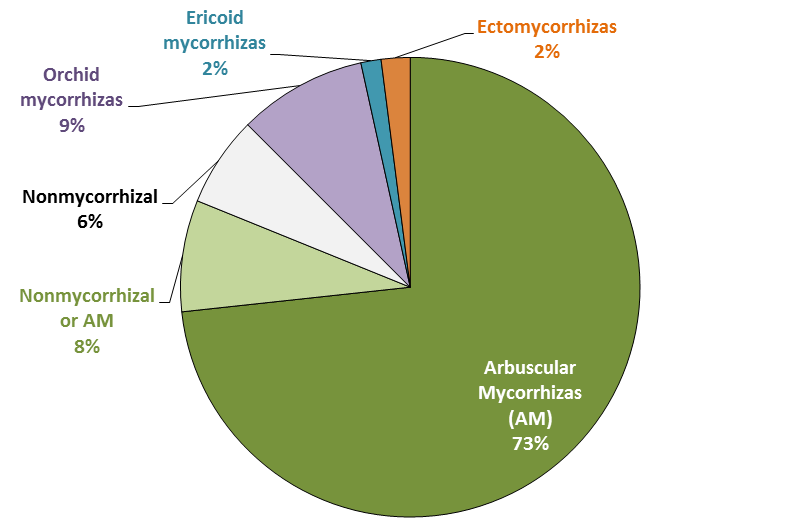
Figure 4.21 Relative importance of mycorrhizal associations for all flowering plants. About 94% of plants can form mycorrhizas of various types. Arbuscular mycorrhizas (AM) are the most common type. Shown in light green are the 8% of species with inconsistent associations that vary with habitat or soil conditions and can be nonmycorrhizal or AM. (Based on Brundrett 2009)
Mycorrhizal fungi are thought to have aided the first plants to colonise land, but most or all species in some plant genera have subsequently lost the ability to form mycorrhizas (e.g. Lupinus, Brassica and Banksia). Nonmycorrhizal plants may have roots that are consistently free of mycorrhizal fungi or have inconsistent associations. The former tend to have alternative nutrition strategies, the latter occur in soils where fungal activity is inhibited, at least part of the time. On a global scale, nonmycorrhizal plants tend to be more common in colder arctic and alpine habitats, and wetland and aquatic habitats, as well as in saline soils and arid habitats (Figure 4.22). These habitats also include many plants with facultative mycorrhizal associations that are present in some cases and not others (called “nonmycorrhizal or AM” in Figure 4.21). In other cases, plants loose the capacity to form mycorrhizas because they are redundant. These include parasites and carnivores, which do not need to acquire nutrients directly from soil (Figure 4.22).
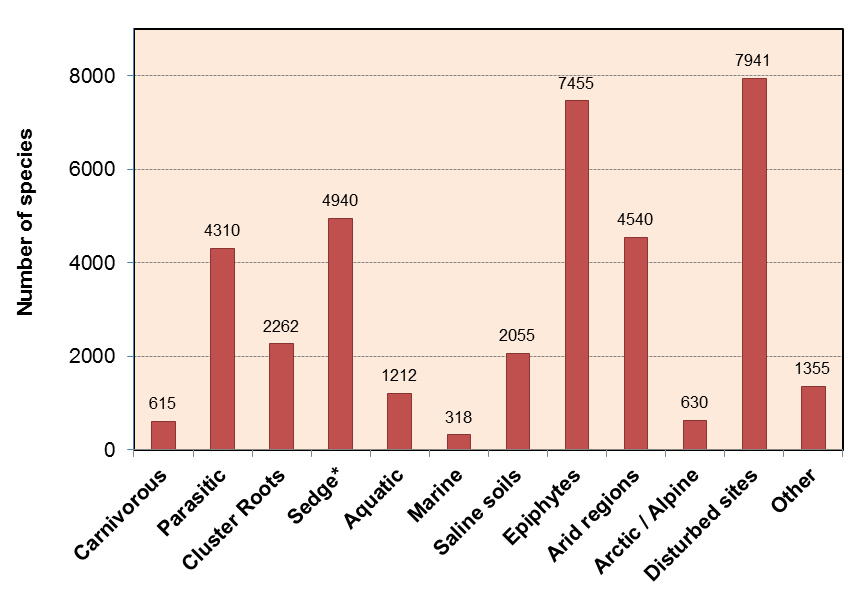
Figure 4.22 Categories and numbers of nonmycorrhizal plants. Most nonmycorrhizal plants occur in specialized habitats where fungal activity is likely to be restricted or have specialized nutritional uptake mechanisms such as carnivory, parasitism or cluster roots. (Data from M. Brundrett, Plant Soil 320: 37-77, 2009)
Mycorrhizal associations are classified according to the way in which the fungi interact with the host plant root, in particular, the structure of the interface that forms between host cells and fungal hyphae. This classification leads to a number of distinct types of mycorrhizal association, as defined in Table 4.1. However, only two of these are widely distributed in the plant kingdom: arbuscular mycorrhizas (AM) and ectomycorrhizas. Orchid and ericoid mycorrhizas are confined to genera within the Orchidaceae and Ericaceae families, respectively.
Mycorrhizal types generally form with a characteristic group of plant species, but there are occasional examples of overlap, such as many Australian plants in the families Fabaceae and Myrtaceae, which have both arbuscular mycorrhizas and ectomycorrhizas. Arbuscular mycorrhizas occur in a vast array of herbaceous genera. In fact, as shown in Figure 4.21 above, some 75% of all plant species form arbuscular mycorrhizas, including most major crop species, that is, all cereals and most grain legumes and pasture legumes.
Table 4.1 shows that the main types of mycorrhizas differ in host preference and in the structures they form during association with the host root, but they are similar in the ways by which they enhance host plant nutrition. Each type of mycorrhiza can be formed by many species of fungi and a single root may often be colonised by more than one species.
Plants with arbuscular mycorrhizas are common in most ecosystems, but are more likely to be dominant in regions of relatively high mean annual temperatures and rates of evapotranspiration, where phosphorus availability is often the major limiting factor for plant growth. However, in some soils with very low phosphorus availability, nonmycorrhizal plant species with cluster roots may be locally dominant and these plants seem to be more efficient at obtaining phosphorus from these soils than mycorrhizal species (e.g. south west Western Australia).
Ectomycorrhizas are most common in tree species, but also occur in some shrubs. In the Northern hemisphere, ectomycorrhizal associations are typically dominant in boreal forests where temperatures and evapotranspiration are relatively low, leading to slow rates of decomposition and accumulation of plant litter in soil and low nitrogen availability. However, ectomycorrhizal plants are also dominant or co-dominant in many other temperate forests, as well as some tropical and subtropical areas, where soil properties are not substantially different from habitats where only arbuscular mycorrhizal plants occur.
Each type of mycorrhizal association has evolved separately to enhance growth and survival of both the host plants and the mycorrhizal fungi. While the primary role of these associations is to increase nutrient supply to the host plant, mycorrhizas have also been shown under some circumstances to enhance plant water status, confer protection against root pathogens, contribute to soil structure through hyphal binding of soil particles and other processes, and render plants less susceptible to toxic elements. The relative importance of these secondary roles is very difficult to determine since they are difficult to separate from nutritional benefits to plants in experiments and they will not be considered in detail here.
During the infection process, fungal hyphae penetrate the epidermal cell layer, often forming distinctive large hyphae within the root at the point of entry (Figure 4.24). From the entry point, hyphae then spread through the root cortex by growing either through the intercellular spaces or from cell to cell by penetrating the cell walls. Hyphae do not, however, penetrate the endodermis or enter the stele.
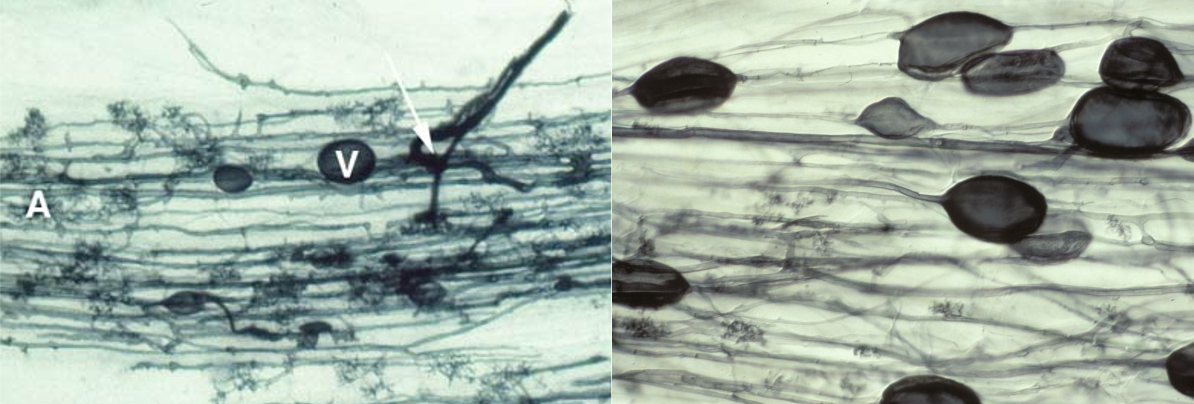
Figure 4.24 Left, Root of clover (Trifolium subterraneum) colonised by an arbuscular mycorrhizal fungus. The fungus has formed thick entry hyphae in the epidermis before spreading through the cortex cells, forming an arbuscule (A) in many cortex cells and some vesicles (V). Roots were cleared (to make them transparent) and then stained with Trypan blue. Right, Root of leek (Allium porrum) colonised by indigenous mycorrhizal fungi showing hyphae, arbuscules and many large vesicles. (Photographs courtesy M. Brundrett)
Within individual cortex cells, hyphae may form a distinctive structure called an arbuscule. From the base of each arbuscule, hyphae repeatedly branch, becoming thinner and thinner as they do so (Figure 4.25). The host cell plasma membrane is never penetrated by the fungus.
Thus in a cell with an arbuscule, the host cell plasma membrane remains intact and functional, but proliferates to surround the arbuscular branches (Figure 4.26). The highly-branched nature of arbuscules is thought to increase the surface area to volume ratio of the host plant plasma membrane by up to 20-fold, relative to unoccupied root cells, thus providing an extensive interface across which nutrient exchange can take place (Figure 4.26).
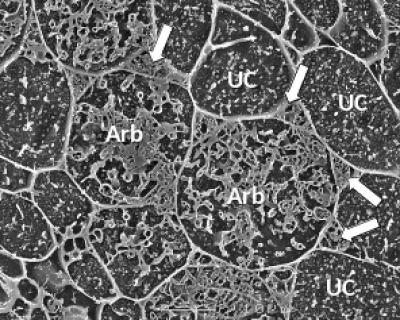
Figure 4.26 A transverse view of cortex cells in frozen roots of white clover (Trifolium repens) colonised by indigenous arbuscular mycorrhizal fungi. In uncolonised cells (UC), the plasma membrane is so closely aligned to the cell wall that it cannot be distinguished and what can be seen inside the cell are dots and lines formed by solutes frozen in the cell vacuole. However, some cells contain the many small hyphae of mature arbuscules (Arb) which have the plasma membrane encasing them and large hyphae are evident occupying most of the intercellular spaces surrounding these cells (arrows). Roots were frozen in liquid nitrogen and viewed using cryo-scanning electron microscopy. (Photograph courtesy M. McCully)
When we consider that all fungal biomass was built using plant carbon, it becomes evident that considerable carbon is needed to maintain the symbiosis.
Arbuscular mycorrhizal fungi store the carbon they obtain from the host plant root primarily in the form of lipids. Lipids are particularly dense in vesicles and spores, which also act as inoculum. Vesicles and spores may form within or outside of roots and often develop most prolifically when roots begin to senesce. The morphology of the fungal infection, particularly the vesicles and spores, differs with the species of fungi.
Ectomycorrhizal symbioses are formed primarily by higher fungi in the Basidiomycotina and Ascomycotina, which form mycorrhizas with the short lateral roots of trees (Table 4.1). Unlike arbuscular mycorrhizas and ericoid mycorrhizas, hyphae of ectomycorrhizal fungi do not normally penetrate host cell walls. Rather, they form an entirely extracellular interface, with highly branched hyphae growing between epidermal or cortical cells, forming a network known as the Hartig net (Figure 4.27).
In Gymnosperms such as Pinus, the Hartig net may extend through most of the root cortex, but in most Angiosperms it is confided to the epidermis (Figure 4.28). In both cases, the highly branched hyphae of the Hartig net provide a substantial surface area for nutrient exchange between the fungus and the plant. Ectomycorrhizas are further differentiated from the other mycorrhizal types by the fact that the fungus usually forms a dense hyphal mantle around each short lateral root, greatly reducing its contact with the soil (e.g. Figure 4.28).
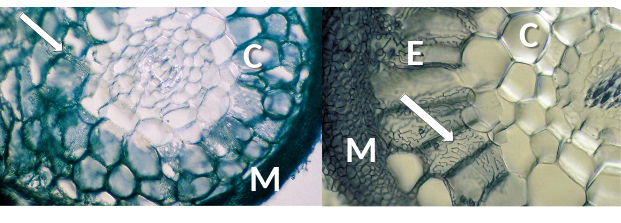
Figure 4.28 Transverse section of ectomycorrhizas showing labyrinthine Hartig net hyphae (arrows) in roots of Pinus sp. (left) and Populus sp. (right). Fungal hyphae are structurally modified, making intimate contact with root cortex (C) (left) or epidermal cells (E) (right) and enabling exchange of resources through the interface between fungus and host. A mantle of fungal hyphae (M) surrounds both roots. (Photographs courtesy M. Brundrett)
Orchid mycorrhizal associations consist of coiling hyphae within cells of a root or stem (Figure 4.29). The most common fungi involved are members of the Rhizoctonia alliance, but ectomycorrhizal fungi are also found in some orchids, especially in achlorophyllous species lacking photosynthesis. A key feature of orchid mycorrhizas is the capacity of fungi to germinate the tiny seeds of orchids to form tiny protocorms which lack roots or leaves. It is thought that orchids start out by exploiting fungi, but then may develop more mutualistic associations as they grow larger and develop leaves.
Ericoid mycorrhizal fungi (largely ascomycetes) form an interface within cells, consisting of dense hyphal coils which are surrounded by host plasma membrane which is similar to orchid mycorrhizas (Figure 4.30). Many members of the Ericaceae host these associations in very fine lateral roots called hair roots, which are only a few cells wide (Figure 4.30).
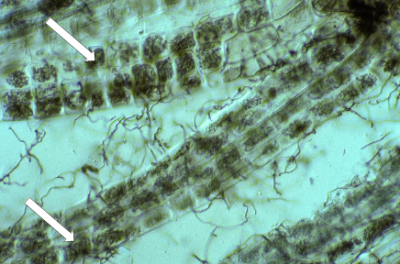
Figure 4.30 Several hair roots of Leucopogon verticillata, an Australian member of the Ericaceae, with nearly every cell containing intracellular hyphal coils of an ericoid mycorrhizal fungi (arrows). Many soil hyphae can also be seen leaving the hair roots. Hair roots stained with Chlorazol black E and viewed with interference contrast. (Photograph courtesy M. Brundrett)
The association between fungus and plant delivers nutrients to the host plant via: (a) mobilisation and absorption by fungal mycelia in the soil; (b) translocation to the fungus–root interface within the root and (c) transfer across the fungus–root interface into the cytoplasm of root cells. As shown in Figure 4.31, both roots and mycorrhizas can absorb nutrients such as phosphorus from the soil, so plants with highly branched fine roots and long root hairs are less likely to benefit substantially from mycorrhizal associations.
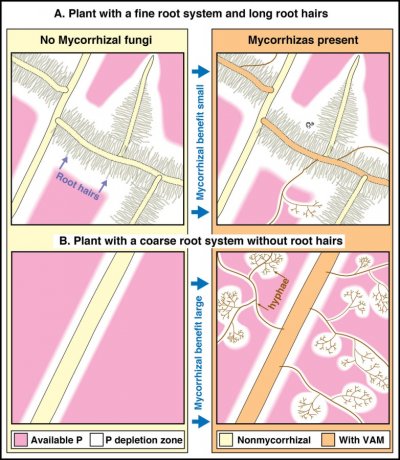
Figure 4.31 Diagrammatic summary showing the impact of roots hairs or arbuscular mycorrhizal fungal hyphae on phosphorus uptake from the soil. Compare the upper and lower pairs of drawings to see how soil hyphae increase the size of phosphorus depletion zones in soil much more if plants lack highly branched roots with long root hairs. (Based on Brundrett et al. 1996)
In addition to hyphae in direct contact with the root surface, all mycorrhizal fungi produce soil hyphae (extramatrical mycelium) which extend into the surrounding soil. Both arbuscular mycorrhizal and ectomycorrhizal fungi can produce copious soil hyphae, that extends well beyond the nutrient depletion zone for immobile nutrients around individual roots and display a complex architecture that renders them an efficient nutrient-collecting and transport network (Figure 4.33). The soil hyphae of many ectomycorrhizal fungi form hyphal aggregations, known as mycelial strands and rhizomorphs, that play a major role in transport of inorganic nutrients or photoassimilates.
Extramatrical mycelium is also primarily responsible for spread of the association to new roots and translocation of energy from the plants for fungal reproduction. Fungi forming mycorrhizal associations can also spread by germination of wind or animal dispersed spores and, in some cases, from old root pieces.
In arbuscular mycorrhizas, fine highly branched soil hyphae (diameter 1–5 µm) provide surface area for nutrient absorption, while larger diameter hyphae (up to 10 µm) form a transport network in the soil for moving solutes from bulk soil to the root (Figure 4.34). Absorption of phosphate by the fungus is maximised by the action of a high-affinity transporter which is expressed only in the soil hyphae of arbuscular mycorrhizal fungi during symbiosis with the plant. The fungi take up inorganic phosphate and quickly convert it to polyphosphate, a macromolecule where the charge of the phosphate ions is balanced by cations including those of potassium and magnesium. Polyphosphate allows phosphorus to be transported to the plant without affecting hyphal osmotic balance. For instance, within the root, concentrations of phosphorus in hyphae may be up to 350 mM, but plant cell vacuoles generally have < 10 mM. Once the polyphosphate reaches an arbuscule in the plant root it is converted back to phosphate and released into the peri-arbuscular space where it is absorbed by the host plant. Active transporters in the host cell plasma membrane maintain a concentration gradient across the plant-fungus interface.
Many experiments have demonstrated a relationship between arbuscular mycorrhizal infection and improved plant phosphorus status, particularly under glasshouse and laboratory conditions (Figure 4.35). Arbuscular mycorrhizal fungi do not appear to have access to sources of soil phosphorus that are otherwise unavailable to nonmycorrhizal roots. Thus, increased plant absorption in the presence of arbuscular mycorrhizal fungi of phosphorus, nitrogen and other macronutrients such as calcium and sulphur, and micronutrients including zinc and copper, seems to primarily reflect the increased absorptive surface of the soil hyphae. However, soil hyphae also provide a conduit for rapid transport of carbon from plants into soil and there is evidence that hyphal exudation may promote breakdown of organic nutrient sources by other microorganisms (see below). Note that the effects of individual species or strains of fungi on plant nutrition will vary, in part due to different morphology of soil hyphae. Under some circumstances, the presence of arbuscular mycorrhizal fungi may decrease plant growth especially in heavily fertilised crop plants.
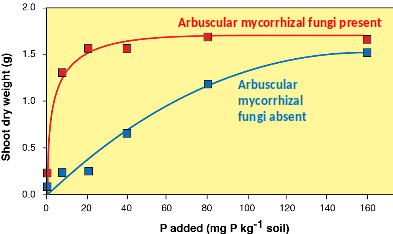
Figure 4.35 The typical growth response for an Australian Cassia species to inoculation with arbuscular mycorrhizal fungi. The greater shoot dry weight of the inoculated plants is due to the fungi enhancing plant uptake of phosphorus. There is no benefit for the plant from the fungi at the lowest level of phosphorus as the fungi are also likely limited by phosphorus, but benefit is substantial at low-intermediate phosphorus levels. (Based on Brundrett et al. 1996, data courtesy of David Jasper and Karen Clarke)
The soil hyphae of ectomycorrhizal fungi increase the absorptive area of a root system substantially, extending the volume of soil explored by the host plant and consequently the quantity of minerals available. Ectomycorrhizal fungi, however, use additional strategies to enhance nutrient acquisition. Some secrete extracellular proteinases, peptidases, phosphomonoesterases and phosphodiesterases that effectively hydrolyse organic nitrogen and phosphorus sources to liberate some nitrogen and phosphorus compounds which can be absorbed by the fungi. Some ectomycorrhizal fungi produce hydrolytic enzymes within the cellulase, hemicellulase and lignase families that may facilitate hyphal entry to moribund plant material in soil and access to mineral nutrients sequestered therein. In these ways, ectomycorrhizal fungi short-circuit conventional nutrient cycles, releasing nutrients from soil organic matter with little or no involvement of saprotrophic organisms. Ectomycorrhizal fungi also release siderophores capable of complexing iron and oxalate to improve potassium uptake and have also been implicated in promoting weathering of rocks to release mineral nutrients for plants.
Arbuscular mycorrhizal fungi depend completely on the host plant for carbon and are unable to grow without being associated with a host plant. This has made the culture of these fungi difficult and proved a significant barrier to development of cheap technologies for inoculation (as might be desirable in land rehabilitation or agriculture) on a large scale. Transfer of carbon from the host to an arbuscular mycorrhizal fungus likely takes place in the arbuscule where the plant releases simple sugars (hexoses) which are absorbed by the fungi. These sugars are rapidly converted into trehalose, glycogen and lipids. The lipids and, to a lesser extent, glycogen are transported to the soil hyphae. Once in the soil hyphae, lipids are progressively broken down into hexoses and trehalose and used to fuel the growth of the fungus. As the lifecycle of the fungus progresses, large amounts of lipid are stored, particularly in vesicles and spores which may be inside or outside of the roots.
In contrast to arbuscular mycorrhizal fungi, ectomycorrhizal fungi can utilise carbon substrates other than those provided by the host plant. It seems that most ectomycorrhizal fungi have some ability to use lignin and cellulose, along with various other substrates including starch, glycogen and sugars such as glucose. Ability to utilise various substrates differs among fungal species. As a result of these abilities, ectomycorrhizal fungi are able to be isolated and grown in culture. For ectomycorrhizal fungi associated with host roots, sucrose is thought to be hydrolysed in root cell walls and glucose to be then absorbed by hyphae from the interface apoplasm.
It has been estimated that 20-50% of plant photosynthate is allocated to mycorrhizal fungi, much of which is allocated to soil hyphae. The soil hyphae of the fungi exude carbon compounds which will influence soil processes including the growth, composition and function of the soil microbial community. Recent research suggests that roots and mycorrhizas may differentially affect soil carbon pools. Thus, overall, the fungi provide a significant pipeline for the movement of carbon from the plant shoot into the soil and may greatly influence soil processes and microbial activity both within and away from the rhizosphere. Indeed, it is now thought that the fungi may significantly influence the global carbon cycle (e.g. their cell walls include some components that are very slow to decompose in soils). In addition, some compounds exuded by the soil hyphae of mycorrhizas, such as glomalin, play an important role in maintenance of soil structure through gluing together soil particles, especially in sandy soils. In addition, colonisation can change the amount and composition of compounds exuded by roots. For instance, the presence of arbuscular mycorrhizal fungi can result in the amount of carboxylates in the rhizosphere being reduced by 50% or more (Figure 4.36). Carboxylates are low molecular weight organic anions which are thought to play a role in release of highly sorbed phosphorus into forms that plant roots or the hyphae of arbuscular mycorrhizal fungi can absorb. Hence, the presence of the fungi enhances the ability of the host to access orthophosphate, but perhaps at the expense of its ability to release phosphorus from sorbed sources.
Overall, there are many fascinating complexities to the relationships among host plants, fungi, nutrients such as phosphorus and nitrogen, carbon and the soil microbial community. For instance, plants make “trade-offs” among nutrient acquisition strategies, probably due to the carbon costs of each strategy. For example, colonisation by arbuscular mycorrhizal fungi often results in a reduction in root / shoot ratio, and root hair production and fine root production tends to be greater in nonmycorrhizal plants. While there are substantial costs to plants in supporting mycorrhizal associations, the cost of producing roots that function well without them seems to be even greater. Overall, the best evidence that mycorrhizal roots are more efficient than nonmycorrhizal roots is provided by the data in Figures 4.21 and 4.22. This global dataset shows that mycorrhizal plants are normally dominant in ecosystems, with the exception of habitats where conditions are likely to suppress fungal activity (e.g. waterlogged, saline, or very cold soils and epiphytic habitats).Brett J Ferguson and Peter M Gresshoff, Centre for Integrative Legume Research, University of Queensland, Australia
Forming nodules and supplying rhizobia with food for nitrogen fixation are energy demanding processes for the host plant. As a result, legumes have developed innate mechanisms to balance their need to acquire nitrogen with their ability to expend energy (reviewed in Ferguson et al. 2010; Reid et al. 2011).
When ample nitrogen is available in the soil, legume plants require fewer nodules to meet their nitrogen demands. Accordingly, they have an inbuilt mechanism in the root to detect the nitrogen content of the surrounding soil. Nitrogen-based compounds, such as nitrate, trigger the production of a small, hormone-like peptide signal, called Nitrate-Induced CLE 1 (NIC1) in soybean. NIC1 is predicted to be perceived by the receptor, Nodule Autoregulation Receptor Kinase (NARK). This perception results in the production of a novel inhibitor signal that acts locally in the root to prevent further nodulation events.
Nodulation is reduced in plants experiencing stress. This likely helps the plant to conserve its resources for combating the stress and for all-important seed development. To date, a number of stress-related factors have been found to inhibit nodule formation locally in the root, including ethylene, salicylic acid and various reactive oxygen species (reviewed in Ferguson and Mathesius 2014). Acidic soil conditions also reduce nodulation, with low pH also causing elevated soil Al3+ levels that negatively affect root growth.
Less than 10% of rhizobia infection events result in the formation of a fully-developed nodule. This is largely due to the Autoregulation Of Nodulation (AON) mechanism that the host plant uses to control its nodule numbers (reviewed by Ferguson et al. 2010; Reid et al. 2011). Mutant legume plants lacking a functional AON pathway are unable to regulate their nodule numbers and as a result exhibit a supernodulation phenotype (i.e. they develop an excessive number of nodules, with up to 25,000 per plant scored, Figure 1). When these supernodulating mutants are induced to form nodules, they are typically reduced in stature and often yield about 20 – 30% less, likely a direct result of their resources being used to form excess nodule structures (Figure 1).

Figure 1 Legume nodulation and autoregulation. A, Soybean plants grown in nitrogen-poor conditions. Wild-Type (WT) plants form functional root nodules when inoculated with compatible, nitrogen-fixing, Bradyrhizobium japonicum. Non-nodulating mutant plants are unable to form nodules (nod-) and supernodulating mutant plants (nod++) are unable to regulate the number of nodules they form. B, Roots of wild-type (WT) and supernodulating mutant (nod++) common bean plants exhibiting mature nodule structures.
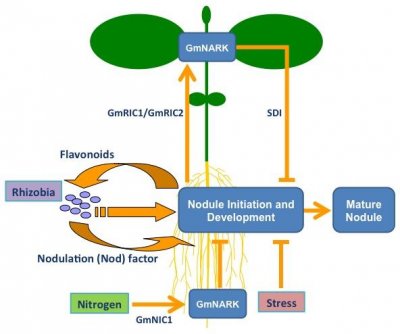
Figure 2 Regulation of legume nodule development. Legume roots exude flavonoid molecules, which attracts compatible rhizobia and triggers them to produce a Nod factor signal. Stress and Nitrogen locally inhibit nodule development. Nitrogen triggers the production of a CLE peptide, called GmNIC1 in soybean, that acts through the GmNARK receptor to suppress nodulation. The Autoregulation Of Nodulation (AON) acts systemically through the shoot. CLE peptides, called GmRIC1 and GmRIC2 in soybean, are produced in the root in response to the first nodulation events. These signals are transported to the shoot where they are perceived by GmNARK, which triggers the production of a shoot-derived inhibitor (SDI) signal that travels to the root to prevent further nodulation.
Although it has not yet been identified, SDI is reported to be a small, heat-stable, Nod factor- and NARK-dependent molecule that is not likely an RNA or protein (Lin et al. 2010) and has been recently been proposed to be the classical phytohormone, cytokinin (Sasaki et al. 2014). Additional factors acting downstream of SDI include Too Much Love, a Kelch-repeat transcription factor whose role in the AON process is yet to be fully defined.
An interesting finding is that of Wang et al. (2014), who suggest that the AON pathway may involve a microRNA after the tentative cytokinin signal. Specifically miR172c was shown to negatively regulate the transcript abundance of a gene (GmNNC1) encoding a transcription factor, being part of the AP2 family. GmNNC1 negatively targets the early nodulin gene ENOD40, needed for nodulation progress in soybean, Medicago truncatula, and Lotus japonicus. It now appears critical to connect the function of a peptide-activated receptor (GmNARK) with the reported cytokinin signal and the subsequent negative regulation cascade described by Wang et al. (2014).
Ferguson BJ, Indrasumunar A, Hayashi S et al. (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52: 61-76
Ferguson BJ, Mathesius U (2014) Phytohormone regulation of legume-rhizobia interactions. J Chem Ecol 40: 770-790
Lin Y-H, Ferguson BJ, Kereszt A, Gresshoff PM (2010) Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent small molecular fraction. New Phytol 185: 1074-1086
Okamoto S, Shinohara H, Mori T et al. (2013) Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nature Comms 4, doi:10.1038/ncomms3191
Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y (2013). Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nature Chem Biol 9: 726-730
Reid DE, Ferguson BJ, Hayashi S et al. (2011) Molecular mechanisms controlling legume autoregulation of nodulation. Ann Botany 108: 789-795
Sasaki T, Suzaki T, Soyano T et al. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nature Comms 5, doi:10.1038/ncomms5983.
Wang Y, Wang L, Zou Y et al. (2014) Soybean miR172c targets the repressive AP2 transcription factor GmNNC1 to activate GmENOD40 expression and regulate nodule initiation. Plant Cell 26: 4782–4801
Ulrike Mathesius, Research School of Biology, Australian National University
Nitrogen is an important nutrient for all plants. While there is an abundance of nitrogen in the atmosphere, plants are unable to convert N2 into a usable form. Fixation of nitrogen gas into ammonia is an ability restricted to nitrogen-fixing bacteria, which contribute most of the inorganic nitrogen to the Earth’s nitrogen cycle.
This chapter explores the importance, evolution and regulation of biological nitrogen fixation, especially of bacteria that have evolved symbiotic associations with higher plant plants. The symbiosis of legumes with nitrogen-fixing soil bacteria called rhizobia has become a model for our understanding of plant-microbe interactions.
Research over the last decade and beyond has revealed major principles and molecular mechanisms of how plants have evolved to recognise their symbiotic partners, how they allow them entry into their root systems, how nutrients are exchanged between the partners and how the symbiosis is controlled systemically to balance demand and supply.
Plant growth is frequently limited by nitrogen. Plants generally obtain nitrogen from soil reserves of nitrate or ammonium (so-called mineral nitrogen) but these reserves are often scarce.
Natural ecosystems can ‘run down’ with respect to nitrogen through soil leaching and fire. Relative abundance of nitrogen-fixing species will then increase. For example, a walk from east to west across Fraser Island, Queensland, will take you across progressively older and more nitrogen deficient sand dunes, and from rainforest to heathland.
Agriculture and horticulture, with its harvest of nitrogen-rich grains or leaves, removes site nitrogen that must be replaced by further mineralisation of soil nitrogen, import of mineral nitrogen (fertiliser) or fixation of atmospheric nitrogen (N2).
The earth’s atmosphere is rich in N2 (about 78% N2) which is very unreactive, due to its stable triple bond. No known eukaryotes have the ability to fix nitrogen, i.e. to reduce atmospheric nitrogen into a usable form like ammonia (NH3). Hydrogen (H2) will react with N2 at very high temperatures and pressures on a catalyst by the Haber-Bosch process, where the pressures needed are 10–100 MPa, the temperatures are 400–550 ºC, and the catalyst is Fe. Then:
\[ N_{2} + 3H_{2} \rightarrow 2NH_{3} \tag{1} \]
Large quantities of ammonia are produced by this method for industrial and agricultural use. Some nitrogen is also fixed in the atmosphere during lightning strikes (Fowler et al. 2013).
Amazingly, some bacteria have the ability to catalyse this reaction with an enzyme complex called nitrogenase donating at least four pairs of electrons to every N2 molecule to effect reduction to two NH4+ and at least one H2. The reaction takes place at ambient conditions, catalysed by the Fe–Mo-containing enzyme, nitrogenase.
\[N_{2} + 16ATP + 8e^– + 10H^+ \rightarrow 2NH_{4}^+ + H_{2} + 16ADP + 6P_{i} \tag{2}\]
Biological N2 fixation is energetically expensive even though it occurs at ambient conditions — estimates fall between 3 and 7 g carbon respired gram of nitrogen fixed (Layzell 1992). Photoassimilate consumed to support N2 fixation is unavailable for other processes such as growth. Consider a crop fertilised with 140 kg N ha–1. An N2 fixer could replace this fertiliser, but only at a cost of at least 420 kg C ha–1. As most plant dry matter contains 40% carbon, this is equivalent to a loss of one tonne of dry matter per hectare! However, in natural ecosystems where no nitrogen fertiliser is applied, and with rising costs of synthetic nitrogen fertiliser, which consumes about 2% of global fossil fuels annually, biological nitrogen fixation confers a distinct advantage to plants that associate with N2-fixing bacteria (Figure 4.39). In fact, our use of nitrogen fertilisers in agriculture has expanded so enormously that pollution of aquatic habitats by nitrogen run-off has moved beyond safe limits for our planet (Steffen et al. 2015). During the last 30 years, research into biological nitrogen fixation, especially in the symbiosis of legumes with rhizobia, has advanced to a point where transfer of this symbiosis to non-legume crops is now becoming a serious goal (Rogers and Oldroyd 2014).
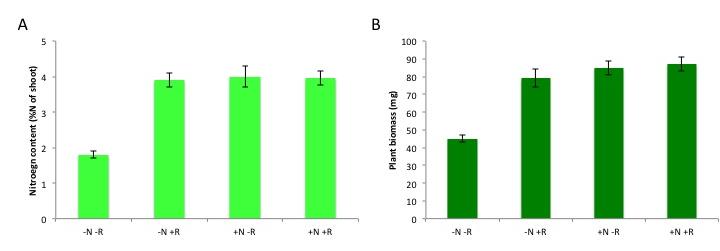
Figure 4.39 Benefit of nitrogen fixation in legumes depending on N fertilisation. Total nitrogen content (A) and plant biomass (B) of subclover (Trifolium subterraneum) plants grown in a glasshouse for four weeks in the presence and absence of nitrogen fertiliser (10 mM potassium nitrate, ‘N’) and/or the rhizobium symbiont (Rhizobium leguminosarum bv. trifolii, ‘R’). Plants benefit from rhizobial inoculation only in the absence of N fertilizer, as shown by the increases in N content and biomass. Rhizobia confer a similar benefit to the unfertilised subclover plants as a 10 mM nitrate addition does, although this varies with different rhizobial strains and with legume species. (Data modified from C.-H. Goh et al., Plant Cell Environ, 2015)
Many bacterial species from diverse phyla have the ability to fix nitrogen, including many (but not all!) cyanobacteria, actinobacteria and proteobacteria. The nitrogen fixation genes are thought to have spread between distantly related bacteria by horizontal gene transfer of clusters of nitrogen-fixation (nif) genes. Nitrogen-fixing bacteria, also called ‘diazotrophs’, can be free-living in water or on solid substrates like soil or rocks. More than half of the biological nitrogen fixation on earth stems from nitrogen-fixing marine bacteria, the rest from terrestrial sources (Fowler et al. 2013). Terrestrial N2-fixing bacteria are found in the soil, in aquatic, and often extreme, habitats, such as hot springs, and nutrient-poor areas. Much of the action of N2-fixing bacteria happens in soils. Plants can then use nitrogen released by decay of such organisms.
However, some plants have evolved a tighter relationship with N2-fixing bacteria, involving an exchange of carbon and nitrogen between plant host and bacterial partner (Santi et al. 2013). Several different symbioses of this type have evolved independently (Table 4.2), from primitive algae to higher vascular plants. These associations are characterised by active attraction of the symbiotic partner through metabolites or signals from the host, as well as a physical housing of the symbiont inside the host (Delaux et al. 2015). For example, algae can form a symbiosis with N2-fixing cyanobacteria to form lichens, and Bryophytes like Anthoceros harbor N2-fixing cyanobacteria in cavities of their thalli. Roots or leaves of some plants form a loose association with N2-fixing bacteria, with plant exudates used as a carbon source by the bacteria. In the water fern Azolla, the cyanobacterium Anabaena is located in cavities on the underside of modified leaves, with a secretory trichome delivering sugars and absorbing fixed nitrogen. This symbiosis is especially important in rice paddy fields, where Azolla densely covers the water surface and can then be incorporated into the field soil. In other plants, the N2 fixers are located in intercellular spaces of the host plant, as reported for sugarcane. These less intimate associations supply host plants with substantial amounts of nitrogen. While such associations have been explored for other grasses, the likely limitation for substantial nitrogen fixation in these loose associations is the limited amount of carbon that the plant provides for its symbionts.In more highly developed associations, plants localise the symbiotic association within a modified root or ‘nodule’. In cycads, the microsymbiont Anabaena is located in intercellular spaces of the mid-cortex of short, highly branched, modified roots (Figure 4.40a and b). In another class of symbioses, the actinorhizal plants, the micro-symbiont Frankia (an actinomycete, or filamentous bacterium) is located within the cortical cells of a modified root. This group includes the genera Casuarina, Allocasuarina, Alnus, Datisca and Myrica from eight plant families, all belonging to the Rosid I class, as do legumes (Figure 4.40c and d). These plants often grow in nutrient-poor habitats where nitrogen fixation provides a nutritional advantage. Parasponia, a tropical tree native in New Guinea, is the only non-legume known to form a symbiosis with the rod-shaped bacterium Rhizobium. Unlike legumes, the Parasponia nodule has a central vascular bundle and the microsymbiont is always encapsulated within cellulosic material (termed a ‘persistent infection thread’). In legumes, nodules typically have a central infected zone, and rhizobia are enveloped by plasma membrane-derived vesicles called symbiosomes (Figure 4.40e and f).
Figure 4.40 Nodule anatomy showing: a, A cycad (Macrozamia miquellii) nodule consisting of a central vascular strand (VB) and an infected cortical region (stars); b, A cyanobacterium, a microsymbiont, is located in the intercellular spaces of this infected cortex; c, A nodule of the river-oak (Casuarina cunninghamii) consisting of a central vascular bundle with infected cells in the cortex (arrows) identified by subersation and lignification of their walls (section stained with berberine sulphate and viewed under epi-fluorescence optics); d, Scanning electron micrograph of an actinomycete microsymbiont (a filamentous bacterium) encapsulated within threads (arrow) throughout the plant cytoplasm; e, A legume (Macroptilium lathyroides) nodule consisting of a central infected region with scattered infected cells (arrows) enclosed in a cortex. Vascular strands (VB) are present in the cortex; f, Transmission electron micrograph of a soybean (Glycine max L.) nodule containing a microsymbiont enveloped by plasma membrane to form ‘symbiosomes’ — packets of bacteria within the cell cytoplasm (arrows). Scale bar = 100 µm in a, b, c and e; 5 µm in d and f. (Images courtesy K. Walsh)
The symbiosis of legumes with rhizobia is the most effective and agriculturally the most important nitrogen-fixing symbiosis. Rhizobia are soil bacteria from the α- and β-proteobacteria (including Burkholderia sp.) that can occur free-living in the soil, but benefit greatly from symbiosis with legume partners, which provide a carbon source, shelter inside a nodule as well as a niche outside the species-rich and competitive zone of the rhizosphere (See previous section on the Soil-root interface). Many species of rhizobia only effectively fix nitrogen inside a nodule (Figure 4.41).
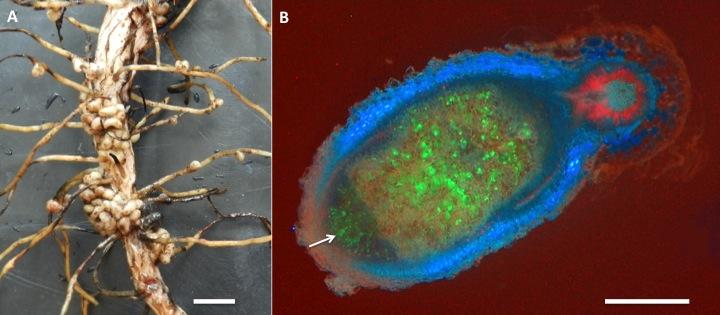
Figure 4.41 Nodules on the root system of a legume. A, Nodules on a root system of a broad bean. B, An infected nodule of Medicago truncatula. Rhizobia express the green fluorescent protein (GFP) and appear in green inside the nodule. New rhizobia infect the tip of the nodule through infection threads (arrow). The blue colour originates from flavonoid autofluorescence in the nodule cortex. Red fluorescence in the main root stems from chlorophyll autofluorescence, as roots were exposed to some light. Magnification bars = 1 cm in A, and 1 mm in B. (Photographs courtesy U. Mathesius)
The ‘precursor’ of symbiosis evolved approximately 100 million years ago, and led to the similar symbioses of actinorhizal plants with Frankia, and the legume symbiosis with rhizobia. Nitrogen fixing symbioses in legumes evolved at a time of relatively high atmospheric CO2. Nitrogen fixation is likely to have conferred an advantage in using this increased CO2 for photosynthesis (Sprent 2007). Phylogenetic reconstruction of symbiotic nitrogen fixation suggests that it evolved once, and was subsequently gained and lost several times in legumes (Figure 4.42; Werner et al. 2014).
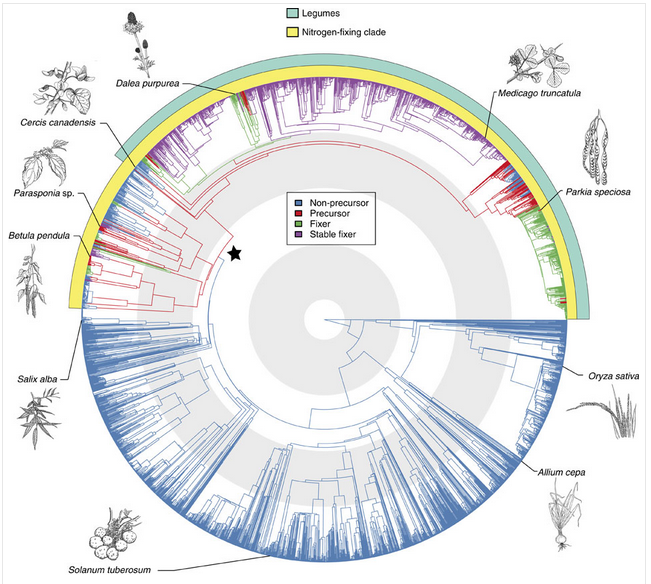
Figure 4.42 Origin of root-nodule symbiosis with rhizobia in angiosperms. Angiosperm phylogeny of 3,467 species showing reconstruction of node states. Branches are coloured according to the most probable state of their ancestral nodes. A star indicates precursor origin. Turquoise and yellow band indicate the legumes and the so-called nitrogen-fixing clade, which contains all known nodulating angiosperms. Grey and white concentric circles indicate periods of 50 million years from the present. The positions of some important angiosperms are indicated with drawings (illustrations by Floortje Bouwkamp). (Reproduced by permission from Macmillan Publishers Ltd from G.D.R. Werner et al. Nature Comms 5: 5087, 2014)
Nodules formed by members of the family Leguminoseae have a central zone of infected cells, surrounded by a cortex of uninfected cells. A root vascular strand branches within the cortex of the nodule. This structure is quite distinct from nodules of the cycads or actinorhizal plants, which have a central vascular bundle and an infected cortex (a typical root vascular anatomy). Different legume species display various nodule growth patterns, but they can be roughly classified as either of indeterminate growth (i.e. with an apical meristem and consequent elongated shape) or determinate growth (i.e. a spherical meristem which ceases activity at nodule maturity).
The association between rhizobia and legumes is a controlled infection. Typically, the bacterial partner infects the plant through root hairs, and is then encapsulated by polysaccharide material produced by the host plant, forming infection threads. Infection threads then grow into the root cortex, while bacteria multiply within each thread. Finally, bacteria are released from the infection threads and engulfed by plant cells in a form of phagocytosis. This process results in a bacterium (sometimes several) encapsulated by a plant cell membrane. Encapsulating membranes control the delivery of photoassimilate to bacteria, thus ensuring a symbiotic rather than a parasitic relationship. These units are termed ‘symbiosomes’ (see also later Section 4.4.5)
Evolution of this partnership might be similar to that of other endosymbiotic organelles such as mitochondria and chloroplasts. Perhaps a future step in the evolution of a legume–rhizobium symbiosis will be retention of bacteria within plant cells to create a new organelle! If this were to happen, the legume would no longer be dependent on the presence of a microsymbiont for infection. Cells could maintain a low resident population of the new organelle, like plastids in non-photosynthetic tissue, and allow proliferation under set conditions within nodule structures.
In some legume symbioses, bacteria are not released from infection threads. This character is one of several that distinguish each of the three legume subfamilies Caesalpinoideae, Mimosoideae and Papilionoideae (e.g. cassia, acacia and soybean, respectively). The Caesalpinoideae are largely trees or shrubs, and the few species which nodulate have little nodule mass proportional to plant biomass (Sprent and Raven 1985). In most of the caesalpinoid species that do nodulate, the microsymbiont remains encapsulated in an infection thread throughout the life of a nodule. In some species the infection threads are thin walled, while in others bacteria are released into the cytoplasm. The Papilionoideae is considered the most advanced of the legume subfamilies.
Biological interactions between host plant and bacterium are subtle. Just as legumes vary genetically, so do the rod-shaped bacteria (rhizobia) that infect various legumes. Not all rhizobia are equally infective (able to infect and form nodules) or effective (able to fix N2) on all legumes. An appropriate bacterial partner must therefore be matched genetically with each legume for optimal N2 fixation. Pure cultures of rhizobia are produced commercially, generally in a peat-moss-based medium or as a seed coating, for inoculating legume seed prior to planting.
Over the last 30 years, the molecular, cellular and genetic analysis of the association between legumes and rhizobia has given us a detailed insight into the specificity and regulation of this symbiosis. As a prerequisite for the symbiosis, the symbionts have to first recognise each other in the soil; the plant host then attracts specific rhizobial symbionts that only nodulate one or a few specific hosts; in response, the rhizobia initiate infection and nodule development. On a whole-plant level, the plant strictly regulates the number of nodules on the root system depending on carbon availability relative to nitrogen demand (See ‘Case Study 4.3 - Regulation of nodule numbers’).
The attraction of rhizobia to the root system of its appropriate host is initiated by the exudation of flavonoids by the plant roots. Flavonoids are a diverse class of phenolic compounds that share a common 15-carbon skeleton consisting of two phenyl rings and a heterocyclic ring, but differ in their exact final structure, for example through modification of the flavonoid ‘backbone’ through hydroxylation, methylation or glycosylation. Over ten thousand different flavonoid molecules have been identified from different plant species. Each legume species exudes a specific mixture of flavonoids into the soil that chemotactically attract their rhizobial symbionts (Figure 4.44). Flavonoids have evolved into specific signalling molecules in the symbiosis by binding specifically to a transcription factor, NodD, inside the rhizobia. This binding of the correct flavonoid activates NodD, and allows it to bind to and activate the promoter regions of a large number of nodulation genes. These nodulation genes of rhizobia are responsible for synthesising a specific signalling molecule, the Nod factor, as well as other genes necessary for symbiosis.
Nod factors are lipochitin-oligosaccharides that are composed of a short ‘backbone’ of N-acetylglucosamine units (the same building blocks that are polymerised to chitin in fungal cell walls, insect exoskeletons and crustacean shells). This backbone typically varies in length between 3-6 units, and is additionally ‘decorated’ with chemical substitutions like acetylation, sulfation or glycosylation (Figure 4.45). Importantly, an acyl side chain of specific length and saturation pattern gives each Nod factor molecule its specificity. Different strains of rhizobia produce specific (mixtures of) Nod factor molecules, which are only recognised by their specific hosts. The Nod factor molecule is crucial for nodulation; mutants defective in Nod factor synthesis cannot nodulate their hosts. Nod factor molecules are also partially sufficient for many of the nodulation steps: application of purified Nod factors to the correct hosts initiates root hair curling and nodule development, but will not lead to infection threads or fully formed nodules. For the complete symbiosis to succeed, other signalling molecules from rhizobia are necessary. For example, surface molecules like exopolysaccharides are necessary to evade host defence responses (Figure 4.45), and communication signals like quorum-sensing signals are necessary to coordinate bacteria behaviours during nitrogen fixation.
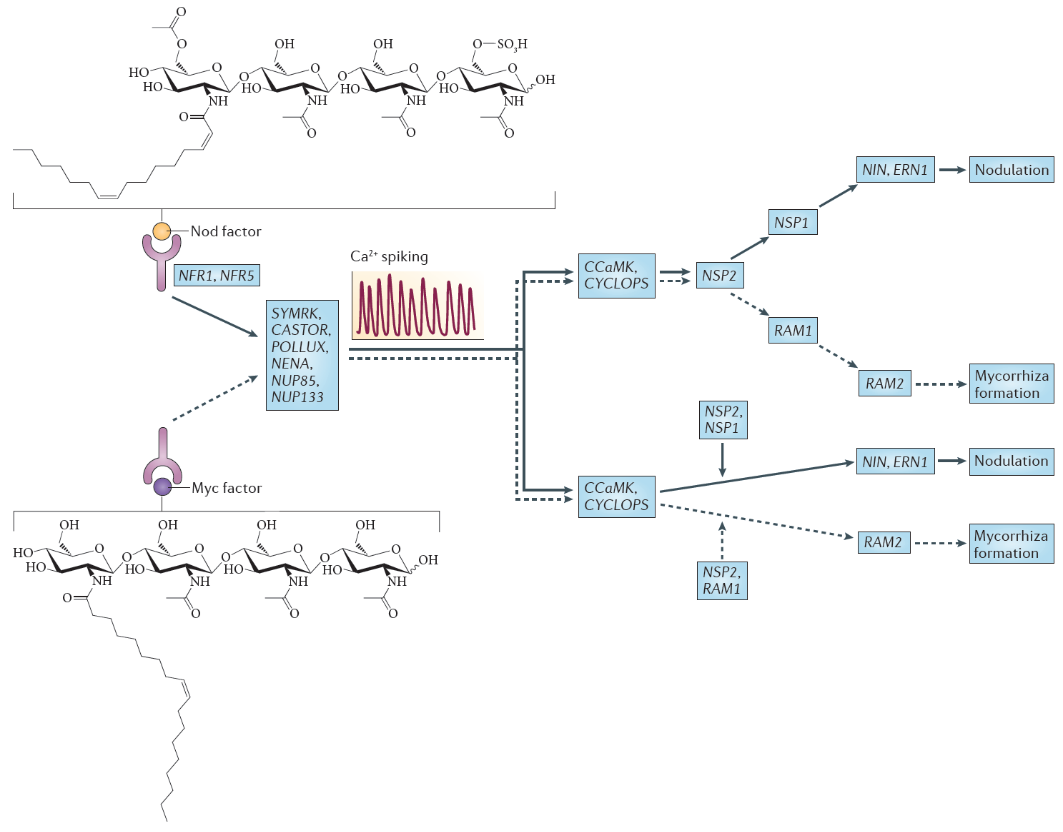
Figure 4.45 Nod factor and Myc factor signalling. The structure of the predominant Nod factor from Sinorhizobium meliloti, the symbiont of the legume Medicago truncatula, is shown (top), in comparison with the structure of the non-sulphated lipochitooligosaccharide Myc factor produced by the arbuscular mycorrhizal fungus Glomus intraradices (bottom). These molecules induce the symbiosis signalling pathway, that shares a calcium spiking response. There are two different interpretations for the genetic overview of this signalling pathway. These interpretations are an amalgamation of genetic analyses in Lotus japonicus and Medicago truncatula; gene names are indicated for the model legume L. japonicus. In the first interpretation (top), the symbiosis signalling pathway transmits the signal through the transcription factors encoded by nodulation signalling pathway 2 (NSP2), NSP1and RAM1 (required for arbuscular mycorrhization 1), whereas in the second interpretation (bottom), symbiosis signalling occurs in parallel to the signalling through these GRAS domain transcription factors. Names for these genes in M. truncatula are as follows: Nod factor receptor 1 (NFR1) is LysM domain receptor kinase 3 (LYK3); NFR5 is Nod factor perception (NFP); symbiosis receptor-like kinase (SYMRK) is DMI2; POLLUX is DMI1; calcium- and calmodulin-dependent serine/threonine protein kinase (CCAMK) is also known as DMI3; CYCLOPS is interacting protein of DMI3 (IPD3). (Reproduced by permission from Macmillan Publishers Ltd from G.E.D. Oldroyd, Nature Rev Microbiol 11: 252-632, 2013)
Nod factors are recognised by their host by Nod factor receptors. Binding of the correct Nod factor structure activates a signalling cascade in the infected root hairs. A critical part of the signalling cascade is a periodic spiking of calcium concentrations in the root hair (‘calcium spiking’), which starts within minutes of Nod factor perception (Figure 4.45). Calcium spiking activates downstream transcription factors that lead to the activation of cytokinin signaling. The plant hormone cytokinin is necessary, and in some cases sufficient to induce cell divisions in the root cortex, the first step of nodule initiation.
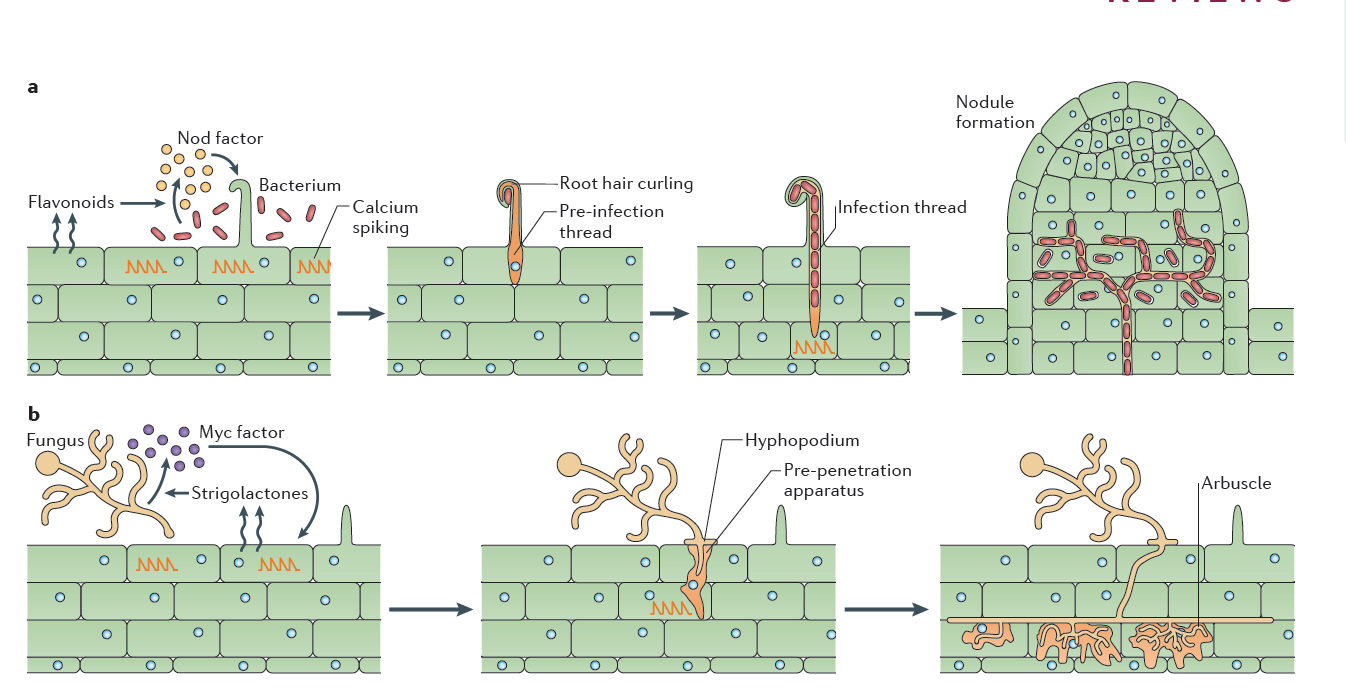
Figure 4.46 Rhizobial and mycorrhizal colonization. a, Flavonoids released by the legume root signal to rhizobia in the rhizosphere, which in turn produce Nod factors that are recognized by the plant. Nod factor perception activates the symbiosis signalling pathway, leading to calcium oscillations, initially in epidermal cells but later also in cortical cells preceding their colonisation (See Figure 3.21). Rhizobia gain entry by infection root hair cells that grow around the bacteria attached at the root surface, trapping the bacteria inside a root hair curl. Infection threads are invaginations of the plant cell that are initiated at the site of root hair curls and allow invasion of the rhizobia into the root tissue. The nucleus relocates to the site of infection, and an alignment of ER and cytoskeleton, known as the pre-infection thread, predicts the path of the infection thread. Nodules initiate below the site of bacterial infection and form by de novo initiation of a nodule meristem in the root cortex. The infection threads grow towards the emergent nodules and ramify within the nodule tissue. In some cases, the rhizobia remain inside the infection threads, but more often, the bacteria are released into membrane-bound compartments inside the cells of the nodule, where the bacteria can differentiate into a nitrogen-fixing state. b, Strigolactone release by the plant root signals to arbuscular mycorrhizal fungi (AMF) in the rhizosphere. Perception of strigolactones promotes spore germination and hyphal branching. AMF produce mycorrhizal factors (Myc factors), including lipochitooligosaccharide (LCOs) and, possibly, signals that activate the symbiosis signalling pathway in the root, leading to calcium oscillations. AMF invasion involves an infection peg from the hyphopodium that allows fungal hyphal growth into the root epidermal cell. The route of hyphal invasion in the plant cell is predicted by a pre-penetration apparatus, which is a clustering of ER and cytoskeleton in a zone of the cell below the first point of fungal contact. The fungus colonizes the plant root cortex through intercellular hyphal growth. Arbuscules are formed in inner root cortical cells from the intercellular hyphae. (Reproduced by permission from Macmillan Publishers Ltd from G.E.D. Oldroyd, Nature Rev Microbiol 11: 252-632, 2013. Part b image is modified by permission from M. Parniske Nature Rev Microbiol 6: 736-775, 2008)
One of the most significant findings of recent years has been the discovery that the same early machinery for Nod factor signal transduction is also required to signal a successful interaction between mycorrhizal fungi and plants (Figure 4.45), and both symbioses share similar mechanisms of bacterial or fungal invasion (Figure 4.46). Mycorrhizal fungi have evolved symbioses with the majority of plant plants much earlier (~400 MYA) than the emergence of the Rhizobium-legume symbiosis (~100 MYA) (see previous section 4.3 on mycorrhiza). Mycorrhizal fungi produce signals that are structurally closely related to Nod factors, and these have been termed ‘Myc factors’. Myc factors are thought to be perceived by different receptors to the Nod factor receptors. Despite some of the shared signals, including calcium spiking, activated by both Myc factors and Nod factors, the later responses in the root differ, leading either to nodulation or to mycorrhization (Figure 4.45). It is still not known what enables legumes to form nodules in response to Nod factors, but other plants not; however, the knowledge that mycorrhizal signalling is part of the response machinery present in all plants that form mycorrhizal symbioses might make it easier to engineer non-legume crops with the ability to form rhizobial symbioses (Rogers and Oldroyd 2014).
Having established the initial contact with the legume host, rhizobia invade the young, just emerging root hairs of their legume partners. In some species, infection can occur intercellularly, and this may have been also the initial mode of infection at the early evolutionary stage of the symbiosis (Held et al. 2014). The infected root hairs curl around small colonies or single cells of rhizobia to form a typical ‘Sheppard’s crook’. The rhizobia locally digest the cell wall of the root hair and induce the formation of ‘infection threads’, tubular invaginations of the root hair in which the rhizobia multiply and travel towards the cortex of the root. This infection step requires correct Nod factor and exopolysaccharide structures from rhizobia, otherwise infection threads are aborted by the plant through defence responses (Figure 4.47). From the infection threads, rhizobia are released into cortical cells as small vesicles, in which rhizobia remain separated from the plant cytoplasm by the plasma membrane. In these vesicles, called symbiosomes, rhizobia differentiate into bacteroids, the nitrogen-fixing forms of rhizobia (Figure 3.49).
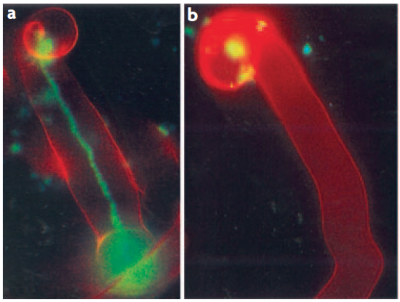
Figure 4.47 Successful and aborted infection threads. a, A successful infection thread of compatible rhizobia on their host. Rhizobia are expressing the Green Fluorescent Protein. b, An aborted infection thread as a result of infection with a rhizobium strain unable to synthesise the correct exopolysaccharides, leading to a defence response. (Reproduced by permission from Macmillan Publishers Ltd from K.M. Jones et al. Nature Rev Microbiol 5: 619-633, 2007)
Concurrent with the growth of the infection thread towards the root cortex, Nod factors produced by the invading rhizobia trigger the re-initiation of cell division in the cortex. These divisions are activated through cytokinin and auxin gradients that the plant forms in response to Nod factors (Desbrosses and Stougaard 2011). In some legumes and actinorhizal plants, the invading rhizobia do not initiate nodules de novo from cortical cells, but target an emerging lateral root primordium that is then ‘converted’ into a nodule.
One surprising recent discovery has been the identification of rhizobia that nodulate legumes in the absence of Nod factors. Sequencing of a large number of Rhizobium species uncovered rhizobial genomes that do not contain any of the canonical nodulation genes that would be required to make Nod factors. It is still unclear what the signal is that these rhizobia use to regulate infection and nodule formation. Similarly, Frankia symbionts do not produce Nod factors, yet use some of the same symbiotic (SYM) signal transduction cascade as legumes. Unravelling the identity and mode of action of these unknown nodulation signals will be a rewarding future challenge.
A basic conflict arises in biological N2 fixation: nitrogenase is destroyed by O2, yet aerobic respiration is essential to sustain the high energy demand of N2 fixation. Nitrogen-fixing bacteria must be protected from O2, while a level of aerobic respiration occurs in the host cell cytoplasm. In cycads, cyanobacteria provide their own O2 protection. Nitrogenase is located in specialised cells (heterocysts) which have an O2-impermeable lining of glycolipid. An analogous structure (a vesicle) affords protection to nitrogenase in the microsymbiont Frankia within most actinorhizal nodules. In Parasponia and most caesalpinoid nodules the persistent infection threads provide O2 protection to nitrogenase (Sprent and Raven 1985).
There is one major problem with structures of ‘fixed’ resistance. As respiration rate varies (i.e. O2 flux), O2 concentration inside the structure must also vary: following Fick’s Law of diffusion, O2 flux into the nodule will change in proportion to the O2 concentration gradient at constant resistance. Free water bathing the nodule is equilibrated with the atmosphere (20.8% O2), therefore containing approximately 360 µM O2 in solution, while nitrogenase is destroyed by submicromolar concentrations of dissolved O2. For practical purposes then, an O2 gradient of 360 (outside) to 0 (inside) µM must be maintained. If respiration rate was halved without a change in resistance, the O2 concentration gradient would also halve from 360 to 180 µM. An O2 concentration of 180 µM O2 inside would destroy nitrogenase. So, resistance must vary too.
A legume-nodule cortex copes with variations in respiration rate by providing a variable level of O2 protection (Layzell and Hunt 1990). According to their model, a layer of cells adjacent to the infected zone either lacks radial intercellular spaces (preventing inflow of O2) or has intercellular spaces filled with water. The thickness of this layer could vary under osmotic control to set nodule permeability. Diffusivity of O2 through water is about 10,000 times slower than through air, so flooding of radial air spaces in the nodule cortex would be an effective way of decreasing O2 diffusion into infected tissue.
Any O2 leaking through this cortex can diffuse freely in the intercellular airspaces of infected tissue and dissolve in the cytoplasm of infected cells. O2 gradients which might be expected within infected cells because of rapid bacterial respiration are largely avoided by the presence of leghemo-globin (Lb) (a molecule similar to the hemoglobin in mammalian blood). O2 diffuses to Lb molecules where it is bound to form high concentrations of oxygenated Lb (estimated at 0.7 mM by Bergersen 1982). Effective nodules are pink because of oxygenated Lb (Figure 4.48); indeed this colour change can be used to estimate free O2 concentrations. Soybean nodules seem to regulate the free O2 in infected cells at between 5–60 nM (e.g. Layzell and Hunt 1990). Finally, residual O2 diffuses through the symbiosome to the bacteroids, supporting a level of aerobic respiration.
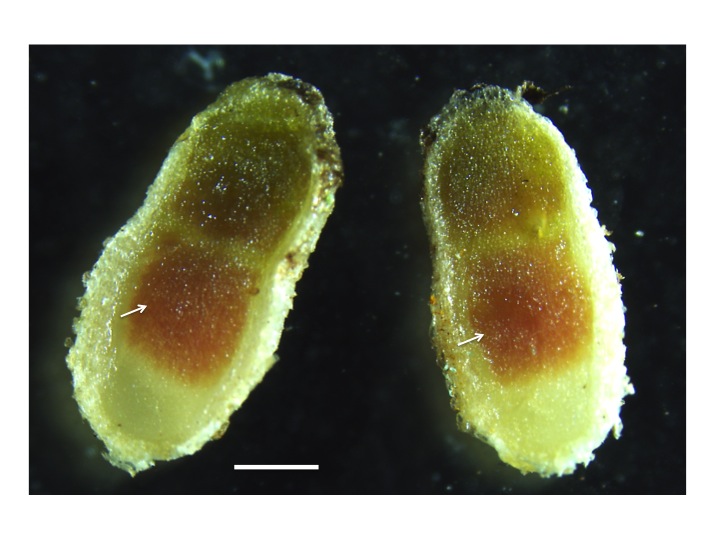
Figure 4.48 Leghemoglobin in nitrogen fixing nodules. The pink colour in the centre of the nodule (arrows) originates from the presence of leghemoglobin in the nitrogen-fixing zone of mature nodules. Leghemoglobin binds oxygen inside the nodules to protect the activity of nitrogenase. It also transports oxygen. Magnification bar = 1 mm. (Photograph courtesy U. Mathesius)
‘Conventional’ chemistry may not be appropriate when describing O2 movement in cells because O2 molecules in cellular compartments are so scarce. A sphere of 1 µm radius — roughly the size of a mitochondrion or bacterium — containing a solution with 10 nM O2 will contain only 24 molecules of O2.
Nodules are metabolically highly active. A typical maximum rate of nitrogenase activity in soybean nodules, as measured by gas exchange (discussed below) is 300 µmol electron pairs g–1 (nodule) h–1. This value is useful to bear in mind when reading the literature about N2 fixation, with low values possibly indicating unhealthy or disturbed plants. As nitrogenase is at best 75% efficient, with respect to N2 fixation (Equation 2), this rate is equivalent to the fixation of some 150 µmol N g–1 (dry weight) h–1. Reduced nitrogen is exported from nodules to the host plant while carbon is imported into the nodule, supporting energy needs of fixation (through respiration) and providing carbon skeletons for packaging nitrogen as an organic molecule.
Photoassimilate (host to nodule) and nitrogen-based resources (nodule to host) must pass through the endodermis of nodule vascular bundles. Radial walls of this endodermis have Casparian bands and tangential walls have relatively few plasmodesmata, so this cell layer restricts apoplasmic and symplasmic flow of carbon into nodules and nitrogen out of nodules.
The transport of solutes, including C and N metabolites but also metal ions, e.g. Fe and Mo required for nitrogenase, are exchanged via both plant and bacterial transporters on the symbiosome and bacterial membrane, and this transport is carefully controlled to balance supply and demand (Figure 4.49; Udvardi and Poole 2013). Not all rhizobia that invade and inhabit legume nodules fix nitrogen efficiently, with the legume host able to restrict C flow to those nodules that do not fix sufficient N.
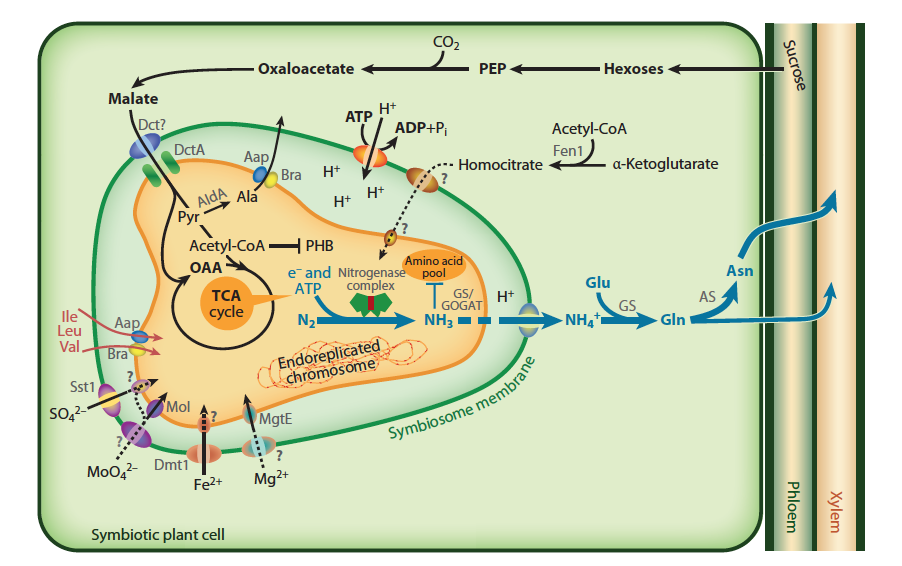
Figure 4.49 Transport and metabolism in an infected nodule cell. Sucrose from the shoot is converted to malate in the plant and imported across the symbiosome membrane and into bacteroids, where it fuels nitrogen fixation. The product of the nitrogen fixation is then exported back to the plant, where it is assimilated into asparagine (Asn) for export to the shoot (blue arrows). In many legumes, such as soybean, the export products are ureides instead of Asn. The plant must provide metals and ions to the bacteroid, although only some of the transport systems on the symbiosome and bacteroid membranes are defined. Many rhizobia lack the ability to make homocitrate or become symbiotic auxotrophs for supply of branched-chain amino acids and become dependent on the plant. (Reproduced with permission by Annual Reviews from M. Udvardi and P.S. Poole, Annu Rev Plant Biol 64: 781-805, 2013)
The concentration of nitrogenous solutes in the xylem apoplasm causes a hydrostatic pressure to develop, and this results in a mass flow of nodule xylem sap to adjacent roots. The water that accompanies sucrose entering the nodule as phloem sap is re-exported with assimilated nitrogen in the xylem. Nodules are thus analogous to ‘glands’ that secrete nitrogenous compounds.
Rates of N2 fixation can be measured by a number of techniques to address questions of nodule efficiency and nitrogen cycling in agricultural and natural plant systems. Nitrogenase is pivotal for initial reduction of N2 but this same enzyme will also reduce acetylene (C2H2) to ethylene (C2H4). Acetylene is an effective competitor with N2 for nitrogenase so the rate of C2H4 synthesis is proportional to nitrogenase activity. Acetylene reduction gives an instantaneous estimate of the N2 fixation rate. Another instantaneous technique requires flushing nodulated roots with an argon : oxygen gas mixture (79:21) to displace all N2. All electron flux through nitrogenase is then diverted to the reduction of protons to H2 rather than N2 to NH4+ (Equation 2). The rate of H2 evolution by roots can thus be used to estimate nitrogenase activity.
Alternative approaches to ‘instantaneous’ estimates of N2 fixation provide an integrated rate of fixation over periods of hours or days. The proportions of inorganic and organic nitrogen compounds in xylem sap are affected by the ratio of inorganic nitrogen taken up to symbiotic N2 fixation; this can be exploited in genera of legumes in which amides and ureides are major products of N2 fixation. Soybean, for example, exports less than 10% of nitrogen to shoots in the form of ureides when supplied nitrate but more than 80% when all nitrogen is biologically fixed. Thus, relative ureide levels in sap give an estimate of N2 fixation.
Many experiments now rely on 15N-based techniques to obtain an integral of fixation over the life of a plant. These techniques rely on a difference in ratio of the stable isotopes of nitrogen (15N and 14N) in soil and atmosphere (Figure 4.50). The soil must be enriched in 15N relative to the atmosphere — either naturally (the process of denitrification causes a fractionation of the two isotopes, leaving the soil enriched in 15N) or by artificial 15N addition. The N2-fixing plant of interest is sampled, together with an adjacent non-N2-fixing plant (e.g. grass) whose 15N enrichment represents that of soil nitrogen. 15N enrichment in digested plant material and soil is analysed isotopically in a mass spectrometer and contribution of biological N2 fixation calculated.
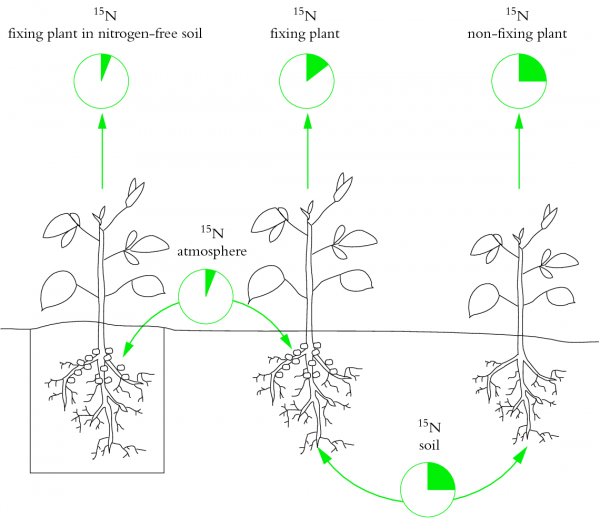
Figure 4.50 Basis of the natural abundance method for assessing the contribution of N2 fixation to legume nutrition. This method entails measuring plant 15N/14N ratio by mass spectrometry. Natural differences in 15N/14N ratio between soil and atmospheric nitrogen are exploited. Legumes to the left and right of the figure each have a unique source of nitrogen, while a test plant in the middle relies on both fixed nitrogen and soil inorganic nitrogen. Plants (left) denied a source of inorganic nitrogen (e.g. nitrate) fix atmospheric nitrogen and therefore have low 15N/14N ratios. Plants without nodules (right) take up only soil-derived nitrogen and are enriched with 15N (high 15N/14N ratios). 15N 'signatures' of these two sets of plants can be used to estimate the relative contributions of soil and atmospheric nitrogen as nitrogen sources in the test plant, and therefore to assess the significance of N2 fixation. (Based on Peoples et al. 1989; reproduced with permission of ACIAR)
A typical ‘good’ rate of fixation for a (non-irrigated) field of subtropical legumes in northern Australia is c. 60–100 kg N ha–1 year–1. About the same amount of nitrogen is harvested as seed from a crop of cowpea, soybean or chickpea, so growing these legumes does not add net nitrogen to the soil; it does, however, spare nitrogen which would otherwise be removed at harvest. Irrigated legume-based pastures in temperate Australia or New Zealand fix 250–300 kg N ha–1 year–1 and make a substantial contribution to the low energy costs of agriculture in these regions. Selection of appropriate biological N2 fixers could greatly improve N2 fixation in tropical legume crops.
Bakker PAHM, Berendsen RL, Doornbos RF et al. (2013) The rhizosphere revisited: root microbiomics. Front Plant Sci 4: Article 65
Barber DA Martin JK (1976) The release of organic substances by cereal roots into soil. New Phytol 76: 69-80
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant Health. Trends Plant Sci 17: 478-486
Bergesen FJ (1982) Root Nodules of Legumes, Structure and Function. Research Studies Press, John Wiley, New York
Blossfeld S, Gansert D, Thiele B et al. (2011) The dynamics of oxygen concentration, pH value, and organic acids in the rhizosphere of Juncus spp. Soil Biol Biochem 43: 1186-1197
Blossfeld S, Schreiber CM, Liebsch G et al. (2013) Quantitative imaging of rhizosphere pH and CO2 dynamics with planar optodes. Ann Bot 112: 267-276
Brundrett MC (2004) Diversity and classification of mycorrhizal associations. Biol Rev 78: 473-495
Brundrett MC (2008) Mycorrhizal Associations: The Web Resource (URL: mycorrhizas.info)
Brundrett MC (2009). Mycorrhizal associations and other means of nutrition of vascular plants: Understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant Soil 320: 37-77
Brundrett M, Bougher N, Dell B et al. (1996) Working with mycorrhizas in forestry and agriculture. Australian Centre for International Agricultural Research. (URL: http://aciar.gov.au/publication/mn032)
Bulgarelli D, Rott M, Schlaeppi K et al. (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91-95
Butler AJ, Barbier N, Čermák J et al. (2010) Estimates and relationships between aboveground and belowground resource exchange surface areas in a Sitka spruce managed forest. Tree Physiol 30: 705-714
Carter EL, Flugga N, Boer JL et al. (2009) Interplay of metal ions and urease. Metallomics 1: 207-221
Delaux P-M, Radhakrishnan G, Oldroyd G (2015) Tracing the evolutionary path to nitrogen-fixing crops. Curr Opin Plant Biol 26: 95-99
Desbrosses GJ, Stougaard J (2011) Root nodulation: A paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10: 348-358
Dittmer HK (1937) A quantitative study of the roots and root hairs of a winter rye plant (Secale cereale). Amer J Bot 24: 417-420
Follmer C (2008) Insights into the role and structure of plant ureases. Phytochem 69: 18-28
Fowler D, Coyle M, Skiba U et al. (2013) The global nitrogen cycle in the twenty-first century. Trans Royal Soc London B 368: 20130164
Goh C-H, Nicotra AB, Mathesius U (2015) The presence of nodules on legume root systems can alter phenotypic plasticity in response to internal nitrogen independent of nitrogen fixation. Plant Cell Environ 39: 883-896
Hartmann A, Rothballer M, Schmid M (2008) Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312: 7–14
Held M, Hossain MS, Yokota K et al. (2010) Common and not so common symbiotic entry. Trends Plant Sci 15: 540-545
Hochholdinger F (2009) The Maize Root System: Morphology, Anatomy, and Genetics. In J.L. Bennetzen and S.C. Hake eds, Handbook of Maize: it’s biology. DOI: 10.1007/978-0-387-79418-1_8
Hochholdinger F, Park WJ, Sauer M, Woll K (2004) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9: 42-48
Jones D, Nguyen C, Finlay DR (2009) Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil 321: 5-33
Jones KM, Kobayashi H, Davies BW et al. (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nature Rev Microbiol 5: 619-633
Jones VAS, Dolan L (2012) The evolution of root hairs and rhizoids. Ann Bot 110: 205-212
Layzell DB (1992) N2 fixation, NO3- reduction and NH4+ assimilation. In Plant Physiology, Biochemistry and Molecular Biology. DT Dennis and DH Turpin, eds, Longman Scientific and Technical: London
Layzell DB, Hunt SH (1990) Oxygen and the regulation of nitrogen fixation in legume nodules. Physiol Plant 80: 322-337
Lundberg DS, Lebeis SL, Herrera Paredes S et al. (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86-90
Malajczuk N, Cromack K (1982) Accumulation of calcium-oxalate in the mantle of ectomycorrhizal roots of Pinus radiata and Eucalyptus marginata. New Phytol 92: 527-531
Marschner H (1995) Mineral nutrition of higher plants (2nd edition) Academic Press, San Diego
Marschner P, Rengel Z (2012) Chapter 12 - Nutrient Availability in Soils. In P Marschner, ed, Marschner's Mineral Nutrition of Higher Plants (Third Edition). Academic Press, San Diego, pp 315-330
McKenzie BM, Mullins CE, Tisdall JM, Bengough AD (2012) Root-soil friction: quantification provides evidence for measurable benefits for manipulation of root-tip traits. Plant Cell Environ 36: 1085-1092
Ohwaki Y Hirata H (1992) Differences in carboxylic acid exudation among P-starved leguminous crops and phospholipid level in roots. Soil Sci Plant Nutr 38: 235-243
Oldroyd GED (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Rev Microbiol 11: 252-632
Parniske M (2008) Arbuscular mycorrhizal: the mother of plant root endosymbioses. Nature Rev Microbiol 6: 736-775
Peoples MB, Faizeh AW, Rerkasem B, Herridge DF (1989) Method for evaluating nitrogen fixation by nodulated legumes in the field. ACIAR Monograph No 11, Canberra
Philippot L, Raaijmakers JM, Lemanceu P, van der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nature Rev Microbiol 11: 789-799
Reich PB (2002) Root-Shoot Relations : Optimality in Acclimation and Adaptation or the "Emperors New Clothes"? In Y Waisel, A Eshel, U Kafkafi, eds, Plant roots: the hidden half. M. Dekker, New York, pp 205-220
Rogers G, Oldroyd GED (2014) Synthetic biology approaches to engineering nitrogen-fixing symbioses in cereals. J Exp Bot 65: 1939-1946
Ryan MH, McCully ME, and Huang CX (2003) Location and quantification of phosphorus and other elements in fully hydrated, soil-grown arbuscular mycorrhizas: a cryo-analytical scanning electron microscopy study. New Phytol 160: 429-441
Ryan MH, Tibbett M, Edmonds-Tibbett T et al. (2012) Carbon trading for phosphorus gain: the balance between rhizosphere carboxylates and arbuscular mycorrhizal symbiosis in plant phosphorus acquisition. Plant Cell Environ 35: 2170-2180
Santi C, Bogusz D, Franche C (2013) Biological nitrogen fixation in non-legume plants. Ann Bot 111: 743–767
Smith SE, Read DJ (2008). Mycorrhizal Symbiosis, Third Edition. Oxford: Academic Press
Sprent JI (2007) Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol 174: 11-25
Sprent JI, Raven JA (1985) Evaluation of nitrogen-fixing symbioses. Proc Royal Soc Edinburgh B 85: 215-237
Steffen W, Richardson K, Rockstroem J et al. (2015) Planetary boundaries: Guiding human development on a changing planet. Science 347: 1259855
Steffens B, Rasmussen A (2016) The physiology of adventitious roots. Plant Physiol 170: 603-617
Teplitski M, Mathesius U, Rumbaugh KB (2011) Quorum sensing signal perception and degradation by mammalian and plant cells. Chem Rev 111: 100-116
Udvardi M, Poole PS (2013) Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64: 781-805
Weaver JE, Himmel WJ (1929) Relation between the development of root system and shoot under long- and short-day illumination. Plant Physiol 4: 435-457
Werner GDR, Cornwell WK, Sprent JI et al. (2014) A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nature Comms 5: 5087