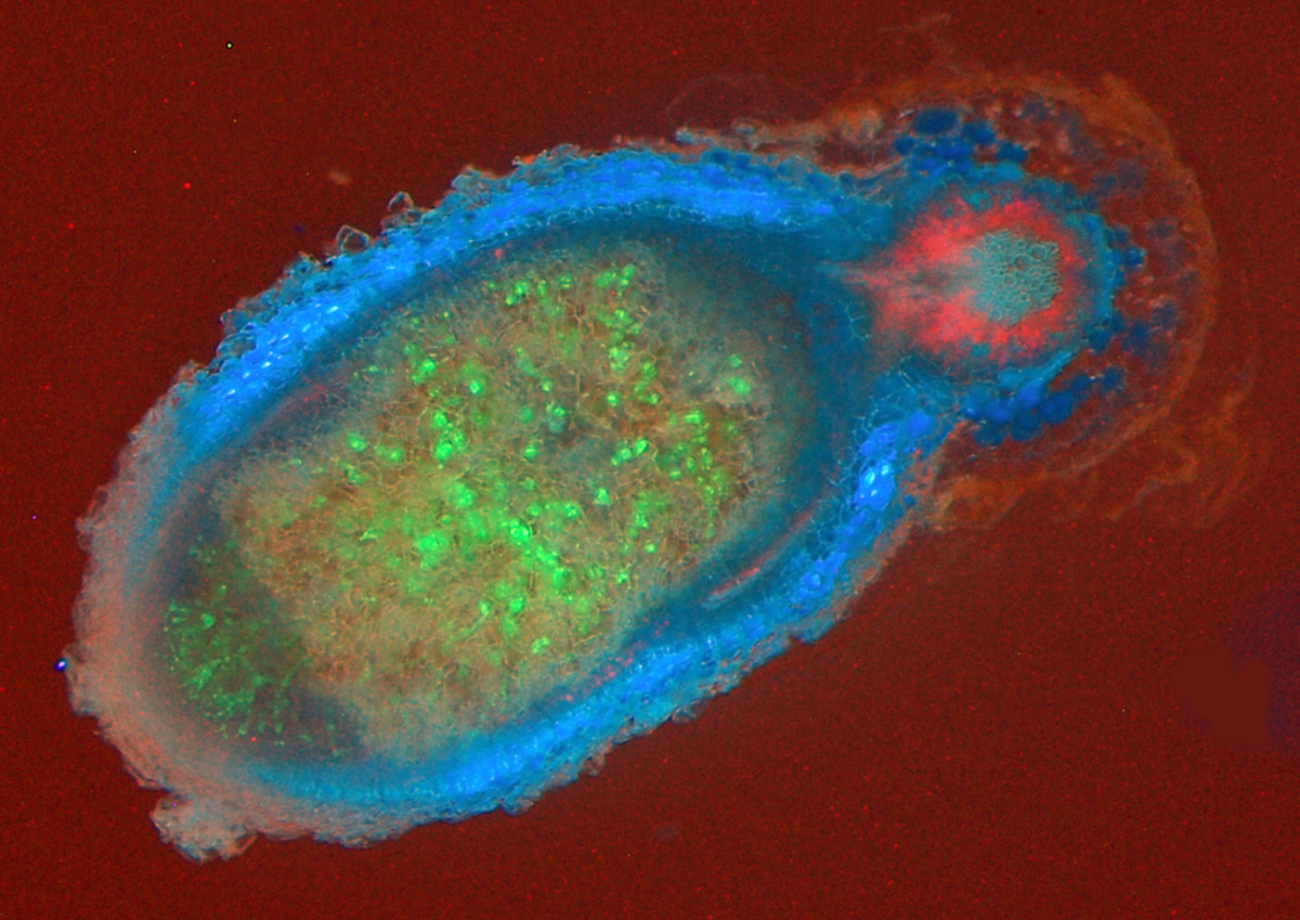
Figure 4.38 An infected nodule of Medicago truncatula. Rhizobia are shown as green inside the large nodule, which is outlined in blue. The root is outlined in red. (Photograph courtesy U. Mathesius)
Ulrike Mathesius, Research School of Biology, Australian National University
Nitrogen is an important nutrient for all plants. While there is an abundance of nitrogen in the atmosphere, plants are unable to convert N2 into a usable form. Fixation of nitrogen gas into ammonia is an ability restricted to nitrogen-fixing bacteria, which contribute most of the inorganic nitrogen to the Earth’s nitrogen cycle.
This chapter explores the importance, evolution and regulation of biological nitrogen fixation, especially of bacteria that have evolved symbiotic associations with higher plant plants. The symbiosis of legumes with nitrogen-fixing soil bacteria called rhizobia has become a model for our understanding of plant-microbe interactions.
Research over the last decade and beyond has revealed major principles and molecular mechanisms of how plants have evolved to recognise their symbiotic partners, how they allow them entry into their root systems, how nutrients are exchanged between the partners and how the symbiosis is controlled systemically to balance demand and supply.
Plant growth is frequently limited by nitrogen. Plants generally obtain nitrogen from soil reserves of nitrate or ammonium (so-called mineral nitrogen) but these reserves are often scarce.
Natural ecosystems can ‘run down’ with respect to nitrogen through soil leaching and fire. Relative abundance of nitrogen-fixing species will then increase. For example, a walk from east to west across Fraser Island, Queensland, will take you across progressively older and more nitrogen deficient sand dunes, and from rainforest to heathland.
Agriculture and horticulture, with its harvest of nitrogen-rich grains or leaves, removes site nitrogen that must be replaced by further mineralisation of soil nitrogen, import of mineral nitrogen (fertiliser) or fixation of atmospheric nitrogen (N2).
The earth’s atmosphere is rich in N2 (about 78% N2) which is very unreactive, due to its stable triple bond. No known eukaryotes have the ability to fix nitrogen, i.e. to reduce atmospheric nitrogen into a usable form like ammonia (NH3). Hydrogen (H2) will react with N2 at very high temperatures and pressures on a catalyst by the Haber-Bosch process, where the pressures needed are 10–100 MPa, the temperatures are 400–550 ºC, and the catalyst is Fe. Then:
\[ N_{2} + 3H_{2} \rightarrow 2NH_{3} \tag{1} \]
Large quantities of ammonia are produced by this method for industrial and agricultural use. Some nitrogen is also fixed in the atmosphere during lightning strikes (Fowler et al. 2013).
Amazingly, some bacteria have the ability to catalyse this reaction with an enzyme complex called nitrogenase donating at least four pairs of electrons to every N2 molecule to effect reduction to two NH4+ and at least one H2. The reaction takes place at ambient conditions, catalysed by the Fe–Mo-containing enzyme, nitrogenase.
\[N_{2} + 16ATP + 8e^– + 10H^+ \rightarrow 2NH_{4}^+ + H_{2} + 16ADP + 6P_{i} \tag{2}\]
Biological N2 fixation is energetically expensive even though it occurs at ambient conditions — estimates fall between 3 and 7 g carbon respired gram of nitrogen fixed (Layzell 1992). Photoassimilate consumed to support N2 fixation is unavailable for other processes such as growth. Consider a crop fertilised with 140 kg N ha–1. An N2 fixer could replace this fertiliser, but only at a cost of at least 420 kg C ha–1. As most plant dry matter contains 40% carbon, this is equivalent to a loss of one tonne of dry matter per hectare! However, in natural ecosystems where no nitrogen fertiliser is applied, and with rising costs of synthetic nitrogen fertiliser, which consumes about 2% of global fossil fuels annually, biological nitrogen fixation confers a distinct advantage to plants that associate with N2-fixing bacteria (Figure 4.39). In fact, our use of nitrogen fertilisers in agriculture has expanded so enormously that pollution of aquatic habitats by nitrogen run-off has moved beyond safe limits for our planet (Steffen et al. 2015). During the last 30 years, research into biological nitrogen fixation, especially in the symbiosis of legumes with rhizobia, has advanced to a point where transfer of this symbiosis to non-legume crops is now becoming a serious goal (Rogers and Oldroyd 2014).
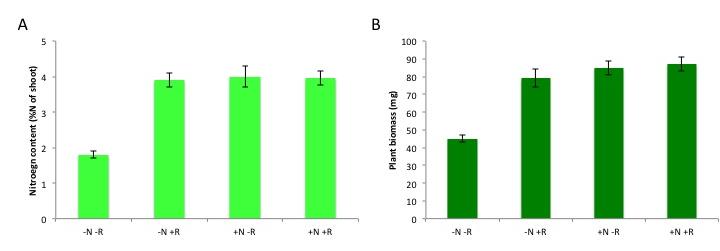
Figure 4.39 Benefit of nitrogen fixation in legumes depending on N fertilisation. Total nitrogen content (A) and plant biomass (B) of subclover (Trifolium subterraneum) plants grown in a glasshouse for four weeks in the presence and absence of nitrogen fertiliser (10 mM potassium nitrate, ‘N’) and/or the rhizobium symbiont (Rhizobium leguminosarum bv. trifolii, ‘R’). Plants benefit from rhizobial inoculation only in the absence of N fertilizer, as shown by the increases in N content and biomass. Rhizobia confer a similar benefit to the unfertilised subclover plants as a 10 mM nitrate addition does, although this varies with different rhizobial strains and with legume species. (Data modified from C.-H. Goh et al., Plant Cell Environ, 2015)
Many bacterial species from diverse phyla have the ability to fix nitrogen, including many (but not all!) cyanobacteria, actinobacteria and proteobacteria. The nitrogen fixation genes are thought to have spread between distantly related bacteria by horizontal gene transfer of clusters of nitrogen-fixation (nif) genes. Nitrogen-fixing bacteria, also called ‘diazotrophs’, can be free-living in water or on solid substrates like soil or rocks. More than half of the biological nitrogen fixation on earth stems from nitrogen-fixing marine bacteria, the rest from terrestrial sources (Fowler et al. 2013). Terrestrial N2-fixing bacteria are found in the soil, in aquatic, and often extreme, habitats, such as hot springs, and nutrient-poor areas. Much of the action of N2-fixing bacteria happens in soils. Plants can then use nitrogen released by decay of such organisms.
However, some plants have evolved a tighter relationship with N2-fixing bacteria, involving an exchange of carbon and nitrogen between plant host and bacterial partner (Santi et al. 2013). Several different symbioses of this type have evolved independently (Table 4.2), from primitive algae to higher vascular plants. These associations are characterised by active attraction of the symbiotic partner through metabolites or signals from the host, as well as a physical housing of the symbiont inside the host (Delaux et al. 2015). For example, algae can form a symbiosis with N2-fixing cyanobacteria to form lichens, and Bryophytes like Anthoceros harbor N2-fixing cyanobacteria in cavities of their thalli. Roots or leaves of some plants form a loose association with N2-fixing bacteria, with plant exudates used as a carbon source by the bacteria. In the water fern Azolla, the cyanobacterium Anabaena is located in cavities on the underside of modified leaves, with a secretory trichome delivering sugars and absorbing fixed nitrogen. This symbiosis is especially important in rice paddy fields, where Azolla densely covers the water surface and can then be incorporated into the field soil. In other plants, the N2 fixers are located in intercellular spaces of the host plant, as reported for sugarcane. These less intimate associations supply host plants with substantial amounts of nitrogen. While such associations have been explored for other grasses, the likely limitation for substantial nitrogen fixation in these loose associations is the limited amount of carbon that the plant provides for its symbionts.In more highly developed associations, plants localise the symbiotic association within a modified root or ‘nodule’. In cycads, the microsymbiont Anabaena is located in intercellular spaces of the mid-cortex of short, highly branched, modified roots (Figure 4.40a and b). In another class of symbioses, the actinorhizal plants, the micro-symbiont Frankia (an actinomycete, or filamentous bacterium) is located within the cortical cells of a modified root. This group includes the genera Casuarina, Allocasuarina, Alnus, Datisca and Myrica from eight plant families, all belonging to the Rosid I class, as do legumes (Figure 4.40c and d). These plants often grow in nutrient-poor habitats where nitrogen fixation provides a nutritional advantage. Parasponia, a tropical tree native in New Guinea, is the only non-legume known to form a symbiosis with the rod-shaped bacterium Rhizobium. Unlike legumes, the Parasponia nodule has a central vascular bundle and the microsymbiont is always encapsulated within cellulosic material (termed a ‘persistent infection thread’). In legumes, nodules typically have a central infected zone, and rhizobia are enveloped by plasma membrane-derived vesicles called symbiosomes (Figure 4.40e and f).
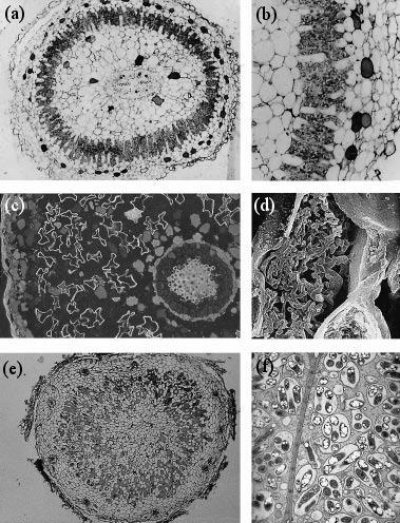
Figure 4.40 Nodule anatomy showing: a, A cycad (Macrozamia miquellii) nodule consisting of a central vascular strand (VB) and an infected cortical region (stars); b, A cyanobacterium, a microsymbiont, is located in the intercellular spaces of this infected cortex; c, A nodule of the river-oak (Casuarina cunninghamii) consisting of a central vascular bundle with infected cells in the cortex (arrows) identified by subersation and lignification of their walls (section stained with berberine sulphate and viewed under epi-fluorescence optics); d, Scanning electron micrograph of an actinomycete microsymbiont (a filamentous bacterium) encapsulated within threads (arrow) throughout the plant cytoplasm; e, A legume (Macroptilium lathyroides) nodule consisting of a central infected region with scattered infected cells (arrows) enclosed in a cortex. Vascular strands (VB) are present in the cortex; f, Transmission electron micrograph of a soybean (Glycine max L.) nodule containing a microsymbiont enveloped by plasma membrane to form ‘symbiosomes’ — packets of bacteria within the cell cytoplasm (arrows). Scale bar = 100 µm in a, b, c and e; 5 µm in d and f. (Images courtesy K. Walsh)
The symbiosis of legumes with rhizobia is the most effective and agriculturally the most important nitrogen-fixing symbiosis. Rhizobia are soil bacteria from the α- and β-proteobacteria (including Burkholderia sp.) that can occur free-living in the soil, but benefit greatly from symbiosis with legume partners, which provide a carbon source, shelter inside a nodule as well as a niche outside the species-rich and competitive zone of the rhizosphere (See previous section on the Soil-root interface). Many species of rhizobia only effectively fix nitrogen inside a nodule (Figure 4.41).
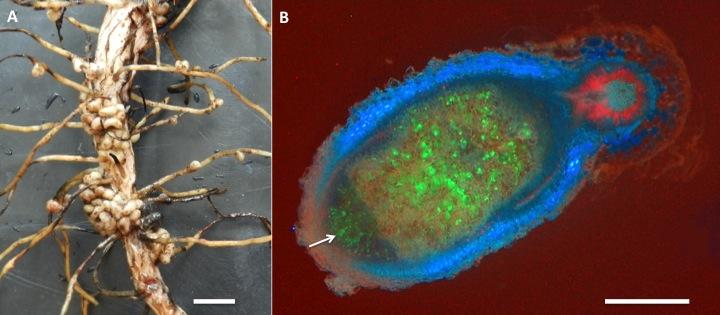
Figure 4.41 Nodules on the root system of a legume. A, Nodules on a root system of a broad bean. B, An infected nodule of Medicago truncatula. Rhizobia express the green fluorescent protein (GFP) and appear in green inside the nodule. New rhizobia infect the tip of the nodule through infection threads (arrow). The blue colour originates from flavonoid autofluorescence in the nodule cortex. Red fluorescence in the main root stems from chlorophyll autofluorescence, as roots were exposed to some light. Magnification bars = 1 cm in A, and 1 mm in B. (Photographs courtesy U. Mathesius)
The ‘precursor’ of symbiosis evolved approximately 100 million years ago, and led to the similar symbioses of actinorhizal plants with Frankia, and the legume symbiosis with rhizobia. Nitrogen fixing symbioses in legumes evolved at a time of relatively high atmospheric CO2. Nitrogen fixation is likely to have conferred an advantage in using this increased CO2 for photosynthesis (Sprent 2007). Phylogenetic reconstruction of symbiotic nitrogen fixation suggests that it evolved once, and was subsequently gained and lost several times in legumes (Figure 4.42; Werner et al. 2014).
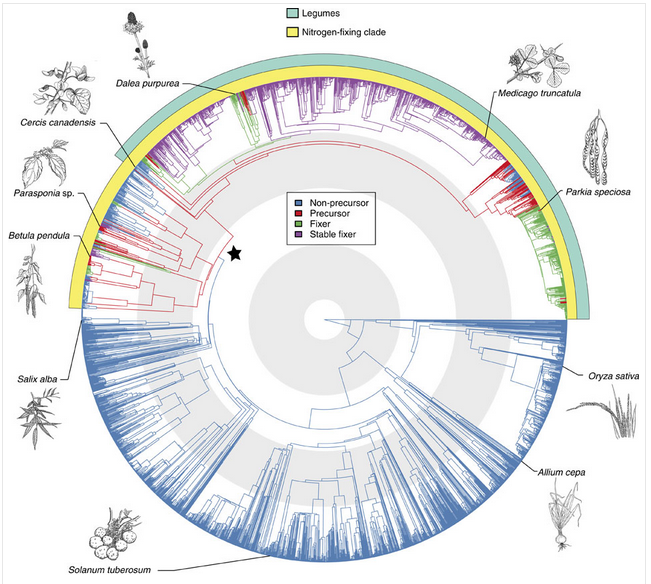
Figure 4.42 Origin of root-nodule symbiosis with rhizobia in angiosperms. Angiosperm phylogeny of 3,467 species showing reconstruction of node states. Branches are coloured according to the most probable state of their ancestral nodes. A star indicates precursor origin. Turquoise and yellow band indicate the legumes and the so-called nitrogen-fixing clade, which contains all known nodulating angiosperms. Grey and white concentric circles indicate periods of 50 million years from the present. The positions of some important angiosperms are indicated with drawings (illustrations by Floortje Bouwkamp). (Reproduced by permission from Macmillan Publishers Ltd from G.D.R. Werner et al. Nature Comms 5: 5087, 2014)
Nodules formed by members of the family Leguminoseae have a central zone of infected cells, surrounded by a cortex of uninfected cells. A root vascular strand branches within the cortex of the nodule. This structure is quite distinct from nodules of the cycads or actinorhizal plants, which have a central vascular bundle and an infected cortex (a typical root vascular anatomy). Different legume species display various nodule growth patterns, but they can be roughly classified as either of indeterminate growth (i.e. with an apical meristem and consequent elongated shape) or determinate growth (i.e. a spherical meristem which ceases activity at nodule maturity).
The association between rhizobia and legumes is a controlled infection. Typically, the bacterial partner infects the plant through root hairs, and is then encapsulated by polysaccharide material produced by the host plant, forming infection threads. Infection threads then grow into the root cortex, while bacteria multiply within each thread. Finally, bacteria are released from the infection threads and engulfed by plant cells in a form of phagocytosis. This process results in a bacterium (sometimes several) encapsulated by a plant cell membrane. Encapsulating membranes control the delivery of photoassimilate to bacteria, thus ensuring a symbiotic rather than a parasitic relationship. These units are termed ‘symbiosomes’ (see also later Section 4.4.5)
Evolution of this partnership might be similar to that of other endosymbiotic organelles such as mitochondria and chloroplasts. Perhaps a future step in the evolution of a legume–rhizobium symbiosis will be retention of bacteria within plant cells to create a new organelle! If this were to happen, the legume would no longer be dependent on the presence of a microsymbiont for infection. Cells could maintain a low resident population of the new organelle, like plastids in non-photosynthetic tissue, and allow proliferation under set conditions within nodule structures.
In some legume symbioses, bacteria are not released from infection threads. This character is one of several that distinguish each of the three legume subfamilies Caesalpinoideae, Mimosoideae and Papilionoideae (e.g. cassia, acacia and soybean, respectively). The Caesalpinoideae are largely trees or shrubs, and the few species which nodulate have little nodule mass proportional to plant biomass (Sprent and Raven 1985). In most of the caesalpinoid species that do nodulate, the microsymbiont remains encapsulated in an infection thread throughout the life of a nodule. In some species the infection threads are thin walled, while in others bacteria are released into the cytoplasm. The Papilionoideae is considered the most advanced of the legume subfamilies.
Biological interactions between host plant and bacterium are subtle. Just as legumes vary genetically, so do the rod-shaped bacteria (rhizobia) that infect various legumes. Not all rhizobia are equally infective (able to infect and form nodules) or effective (able to fix N2) on all legumes. An appropriate bacterial partner must therefore be matched genetically with each legume for optimal N2 fixation. Pure cultures of rhizobia are produced commercially, generally in a peat-moss-based medium or as a seed coating, for inoculating legume seed prior to planting.
Over the last 30 years, the molecular, cellular and genetic analysis of the association between legumes and rhizobia has given us a detailed insight into the specificity and regulation of this symbiosis. As a prerequisite for the symbiosis, the symbionts have to first recognise each other in the soil; the plant host then attracts specific rhizobial symbionts that only nodulate one or a few specific hosts; in response, the rhizobia initiate infection and nodule development. On a whole-plant level, the plant strictly regulates the number of nodules on the root system depending on carbon availability relative to nitrogen demand (See ‘Case Study 4.3 - Regulation of nodule numbers’).
The attraction of rhizobia to the root system of its appropriate host is initiated by the exudation of flavonoids by the plant roots. Flavonoids are a diverse class of phenolic compounds that share a common 15-carbon skeleton consisting of two phenyl rings and a heterocyclic ring, but differ in their exact final structure, for example through modification of the flavonoid ‘backbone’ through hydroxylation, methylation or glycosylation. Over ten thousand different flavonoid molecules have been identified from different plant species. Each legume species exudes a specific mixture of flavonoids into the soil that chemotactically attract their rhizobial symbionts (Figure 4.44). Flavonoids have evolved into specific signalling molecules in the symbiosis by binding specifically to a transcription factor, NodD, inside the rhizobia. This binding of the correct flavonoid activates NodD, and allows it to bind to and activate the promoter regions of a large number of nodulation genes. These nodulation genes of rhizobia are responsible for synthesising a specific signalling molecule, the Nod factor, as well as other genes necessary for symbiosis.
Nod factors are lipochitin-oligosaccharides that are composed of a short ‘backbone’ of N-acetylglucosamine units (the same building blocks that are polymerised to chitin in fungal cell walls, insect exoskeletons and crustacean shells). This backbone typically varies in length between 3-6 units, and is additionally ‘decorated’ with chemical substitutions like acetylation, sulfation or glycosylation (Figure 4.45). Importantly, an acyl side chain of specific length and saturation pattern gives each Nod factor molecule its specificity. Different strains of rhizobia produce specific (mixtures of) Nod factor molecules, which are only recognised by their specific hosts. The Nod factor molecule is crucial for nodulation; mutants defective in Nod factor synthesis cannot nodulate their hosts. Nod factor molecules are also partially sufficient for many of the nodulation steps: application of purified Nod factors to the correct hosts initiates root hair curling and nodule development, but will not lead to infection threads or fully formed nodules. For the complete symbiosis to succeed, other signalling molecules from rhizobia are necessary. For example, surface molecules like exopolysaccharides are necessary to evade host defence responses (Figure 4.45), and communication signals like quorum-sensing signals are necessary to coordinate bacteria behaviours during nitrogen fixation.
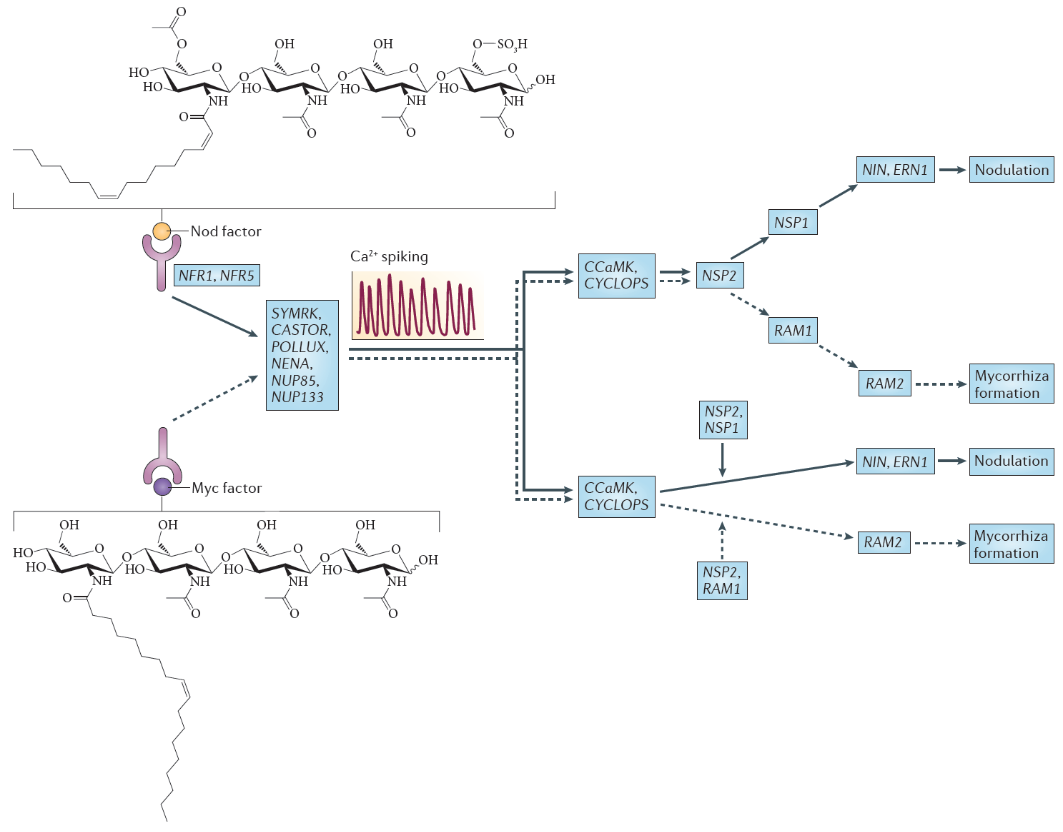
Figure 4.45 Nod factor and Myc factor signalling. The structure of the predominant Nod factor from Sinorhizobium meliloti, the symbiont of the legume Medicago truncatula, is shown (top), in comparison with the structure of the non-sulphated lipochitooligosaccharide Myc factor produced by the arbuscular mycorrhizal fungus Glomus intraradices (bottom). These molecules induce the symbiosis signalling pathway, that shares a calcium spiking response. There are two different interpretations for the genetic overview of this signalling pathway. These interpretations are an amalgamation of genetic analyses in Lotus japonicus and Medicago truncatula; gene names are indicated for the model legume L. japonicus. In the first interpretation (top), the symbiosis signalling pathway transmits the signal through the transcription factors encoded by nodulation signalling pathway 2 (NSP2), NSP1and RAM1 (required for arbuscular mycorrhization 1), whereas in the second interpretation (bottom), symbiosis signalling occurs in parallel to the signalling through these GRAS domain transcription factors. Names for these genes in M. truncatula are as follows: Nod factor receptor 1 (NFR1) is LysM domain receptor kinase 3 (LYK3); NFR5 is Nod factor perception (NFP); symbiosis receptor-like kinase (SYMRK) is DMI2; POLLUX is DMI1; calcium- and calmodulin-dependent serine/threonine protein kinase (CCAMK) is also known as DMI3; CYCLOPS is interacting protein of DMI3 (IPD3). (Reproduced by permission from Macmillan Publishers Ltd from G.E.D. Oldroyd, Nature Rev Microbiol 11: 252-632, 2013)
Nod factors are recognised by their host by Nod factor receptors. Binding of the correct Nod factor structure activates a signalling cascade in the infected root hairs. A critical part of the signalling cascade is a periodic spiking of calcium concentrations in the root hair (‘calcium spiking’), which starts within minutes of Nod factor perception (Figure 4.45). Calcium spiking activates downstream transcription factors that lead to the activation of cytokinin signaling. The plant hormone cytokinin is necessary, and in some cases sufficient to induce cell divisions in the root cortex, the first step of nodule initiation.
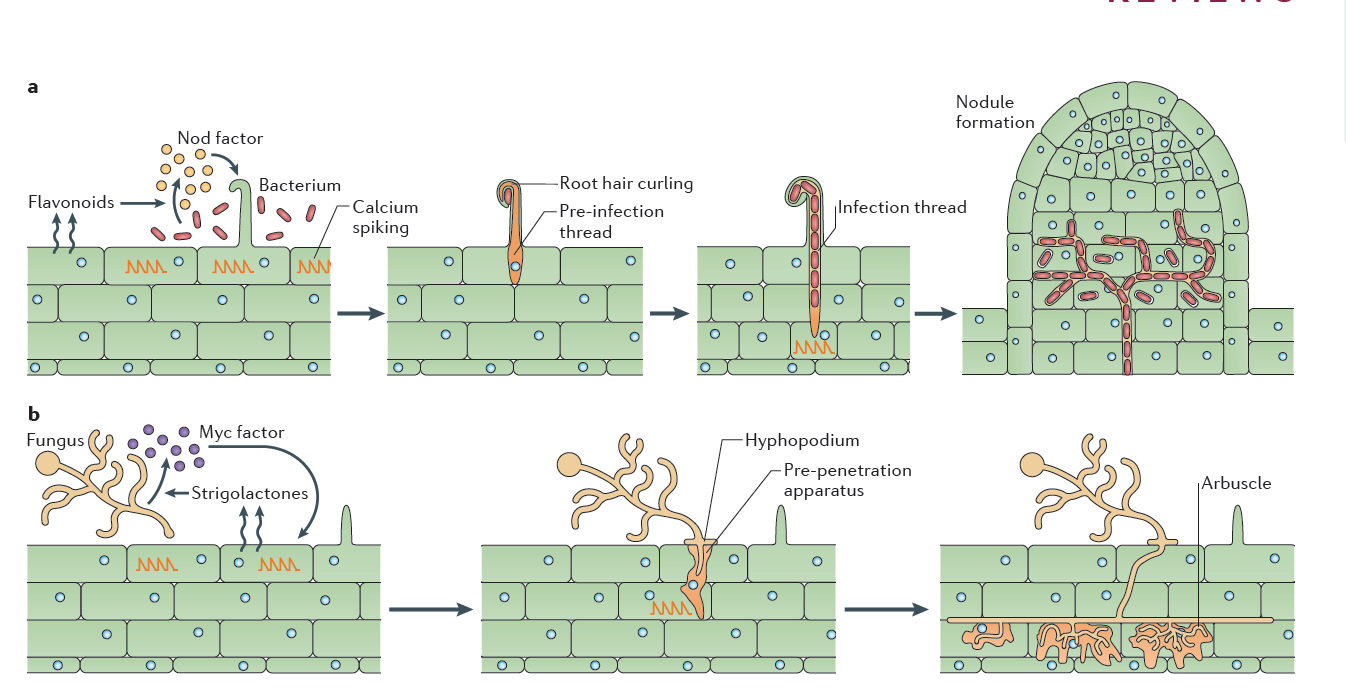
Figure 4.46 Rhizobial and mycorrhizal colonization. a, Flavonoids released by the legume root signal to rhizobia in the rhizosphere, which in turn produce Nod factors that are recognized by the plant. Nod factor perception activates the symbiosis signalling pathway, leading to calcium oscillations, initially in epidermal cells but later also in cortical cells preceding their colonisation (See Figure 3.21). Rhizobia gain entry by infection root hair cells that grow around the bacteria attached at the root surface, trapping the bacteria inside a root hair curl. Infection threads are invaginations of the plant cell that are initiated at the site of root hair curls and allow invasion of the rhizobia into the root tissue. The nucleus relocates to the site of infection, and an alignment of ER and cytoskeleton, known as the pre-infection thread, predicts the path of the infection thread. Nodules initiate below the site of bacterial infection and form by de novo initiation of a nodule meristem in the root cortex. The infection threads grow towards the emergent nodules and ramify within the nodule tissue. In some cases, the rhizobia remain inside the infection threads, but more often, the bacteria are released into membrane-bound compartments inside the cells of the nodule, where the bacteria can differentiate into a nitrogen-fixing state. b, Strigolactone release by the plant root signals to arbuscular mycorrhizal fungi (AMF) in the rhizosphere. Perception of strigolactones promotes spore germination and hyphal branching. AMF produce mycorrhizal factors (Myc factors), including lipochitooligosaccharide (LCOs) and, possibly, signals that activate the symbiosis signalling pathway in the root, leading to calcium oscillations. AMF invasion involves an infection peg from the hyphopodium that allows fungal hyphal growth into the root epidermal cell. The route of hyphal invasion in the plant cell is predicted by a pre-penetration apparatus, which is a clustering of ER and cytoskeleton in a zone of the cell below the first point of fungal contact. The fungus colonizes the plant root cortex through intercellular hyphal growth. Arbuscules are formed in inner root cortical cells from the intercellular hyphae. (Reproduced by permission from Macmillan Publishers Ltd from G.E.D. Oldroyd, Nature Rev Microbiol 11: 252-632, 2013. Part b image is modified by permission from M. Parniske Nature Rev Microbiol 6: 736-775, 2008)
One of the most significant findings of recent years has been the discovery that the same early machinery for Nod factor signal transduction is also required to signal a successful interaction between mycorrhizal fungi and plants (Figure 4.45), and both symbioses share similar mechanisms of bacterial or fungal invasion (Figure 4.46). Mycorrhizal fungi have evolved symbioses with the majority of plant plants much earlier (~400 MYA) than the emergence of the Rhizobium-legume symbiosis (~100 MYA) (see previous section 4.3 on mycorrhiza). Mycorrhizal fungi produce signals that are structurally closely related to Nod factors, and these have been termed ‘Myc factors’. Myc factors are thought to be perceived by different receptors to the Nod factor receptors. Despite some of the shared signals, including calcium spiking, activated by both Myc factors and Nod factors, the later responses in the root differ, leading either to nodulation or to mycorrhization (Figure 4.45). It is still not known what enables legumes to form nodules in response to Nod factors, but other plants not; however, the knowledge that mycorrhizal signalling is part of the response machinery present in all plants that form mycorrhizal symbioses might make it easier to engineer non-legume crops with the ability to form rhizobial symbioses (Rogers and Oldroyd 2014).
Having established the initial contact with the legume host, rhizobia invade the young, just emerging root hairs of their legume partners. In some species, infection can occur intercellularly, and this may have been also the initial mode of infection at the early evolutionary stage of the symbiosis (Held et al. 2014). The infected root hairs curl around small colonies or single cells of rhizobia to form a typical ‘Sheppard’s crook’. The rhizobia locally digest the cell wall of the root hair and induce the formation of ‘infection threads’, tubular invaginations of the root hair in which the rhizobia multiply and travel towards the cortex of the root. This infection step requires correct Nod factor and exopolysaccharide structures from rhizobia, otherwise infection threads are aborted by the plant through defence responses (Figure 4.47). From the infection threads, rhizobia are released into cortical cells as small vesicles, in which rhizobia remain separated from the plant cytoplasm by the plasma membrane. In these vesicles, called symbiosomes, rhizobia differentiate into bacteroids, the nitrogen-fixing forms of rhizobia (Figure 3.49).
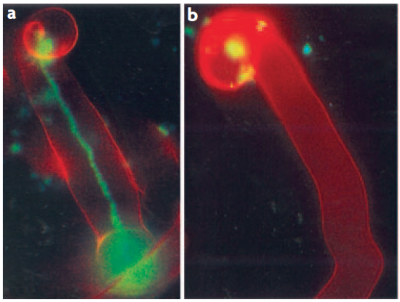
Figure 4.47 Successful and aborted infection threads. a, A successful infection thread of compatible rhizobia on their host. Rhizobia are expressing the Green Fluorescent Protein. b, An aborted infection thread as a result of infection with a rhizobium strain unable to synthesise the correct exopolysaccharides, leading to a defence response. (Reproduced by permission from Macmillan Publishers Ltd from K.M. Jones et al. Nature Rev Microbiol 5: 619-633, 2007)
Concurrent with the growth of the infection thread towards the root cortex, Nod factors produced by the invading rhizobia trigger the re-initiation of cell division in the cortex. These divisions are activated through cytokinin and auxin gradients that the plant forms in response to Nod factors (Desbrosses and Stougaard 2011). In some legumes and actinorhizal plants, the invading rhizobia do not initiate nodules de novo from cortical cells, but target an emerging lateral root primordium that is then ‘converted’ into a nodule.
One surprising recent discovery has been the identification of rhizobia that nodulate legumes in the absence of Nod factors. Sequencing of a large number of Rhizobium species uncovered rhizobial genomes that do not contain any of the canonical nodulation genes that would be required to make Nod factors. It is still unclear what the signal is that these rhizobia use to regulate infection and nodule formation. Similarly, Frankia symbionts do not produce Nod factors, yet use some of the same symbiotic (SYM) signal transduction cascade as legumes. Unravelling the identity and mode of action of these unknown nodulation signals will be a rewarding future challenge.
A basic conflict arises in biological N2 fixation: nitrogenase is destroyed by O2, yet aerobic respiration is essential to sustain the high energy demand of N2 fixation. Nitrogen-fixing bacteria must be protected from O2, while a level of aerobic respiration occurs in the host cell cytoplasm. In cycads, cyanobacteria provide their own O2 protection. Nitrogenase is located in specialised cells (heterocysts) which have an O2-impermeable lining of glycolipid. An analogous structure (a vesicle) affords protection to nitrogenase in the microsymbiont Frankia within most actinorhizal nodules. In Parasponia and most caesalpinoid nodules the persistent infection threads provide O2 protection to nitrogenase (Sprent and Raven 1985).
There is one major problem with structures of ‘fixed’ resistance. As respiration rate varies (i.e. O2 flux), O2 concentration inside the structure must also vary: following Fick’s Law of diffusion, O2 flux into the nodule will change in proportion to the O2 concentration gradient at constant resistance. Free water bathing the nodule is equilibrated with the atmosphere (20.8% O2), therefore containing approximately 360 µM O2 in solution, while nitrogenase is destroyed by submicromolar concentrations of dissolved O2. For practical purposes then, an O2 gradient of 360 (outside) to 0 (inside) µM must be maintained. If respiration rate was halved without a change in resistance, the O2 concentration gradient would also halve from 360 to 180 µM. An O2 concentration of 180 µM O2 inside would destroy nitrogenase. So, resistance must vary too.
A legume-nodule cortex copes with variations in respiration rate by providing a variable level of O2 protection (Layzell and Hunt 1990). According to their model, a layer of cells adjacent to the infected zone either lacks radial intercellular spaces (preventing inflow of O2) or has intercellular spaces filled with water. The thickness of this layer could vary under osmotic control to set nodule permeability. Diffusivity of O2 through water is about 10,000 times slower than through air, so flooding of radial air spaces in the nodule cortex would be an effective way of decreasing O2 diffusion into infected tissue.
Any O2 leaking through this cortex can diffuse freely in the intercellular airspaces of infected tissue and dissolve in the cytoplasm of infected cells. O2 gradients which might be expected within infected cells because of rapid bacterial respiration are largely avoided by the presence of leghemo-globin (Lb) (a molecule similar to the hemoglobin in mammalian blood). O2 diffuses to Lb molecules where it is bound to form high concentrations of oxygenated Lb (estimated at 0.7 mM by Bergersen 1982). Effective nodules are pink because of oxygenated Lb (Figure 4.48); indeed this colour change can be used to estimate free O2 concentrations. Soybean nodules seem to regulate the free O2 in infected cells at between 5–60 nM (e.g. Layzell and Hunt 1990). Finally, residual O2 diffuses through the symbiosome to the bacteroids, supporting a level of aerobic respiration.
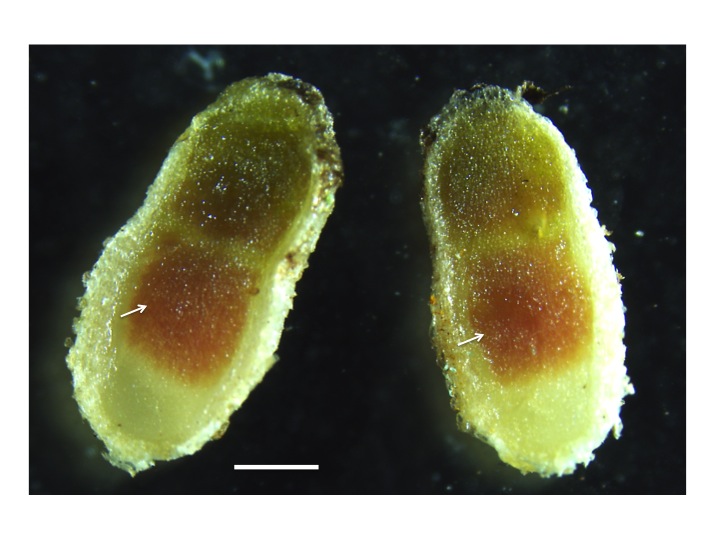
Figure 4.48 Leghemoglobin in nitrogen fixing nodules. The pink colour in the centre of the nodule (arrows) originates from the presence of leghemoglobin in the nitrogen-fixing zone of mature nodules. Leghemoglobin binds oxygen inside the nodules to protect the activity of nitrogenase. It also transports oxygen. Magnification bar = 1 mm. (Photograph courtesy U. Mathesius)
‘Conventional’ chemistry may not be appropriate when describing O2 movement in cells because O2 molecules in cellular compartments are so scarce. A sphere of 1 µm radius — roughly the size of a mitochondrion or bacterium — containing a solution with 10 nM O2 will contain only 24 molecules of O2.
Nodules are metabolically highly active. A typical maximum rate of nitrogenase activity in soybean nodules, as measured by gas exchange (discussed below) is 300 µmol electron pairs g–1 (nodule) h–1. This value is useful to bear in mind when reading the literature about N2 fixation, with low values possibly indicating unhealthy or disturbed plants. As nitrogenase is at best 75% efficient, with respect to N2 fixation (Equation 2), this rate is equivalent to the fixation of some 150 µmol N g–1 (dry weight) h–1. Reduced nitrogen is exported from nodules to the host plant while carbon is imported into the nodule, supporting energy needs of fixation (through respiration) and providing carbon skeletons for packaging nitrogen as an organic molecule.
Photoassimilate (host to nodule) and nitrogen-based resources (nodule to host) must pass through the endodermis of nodule vascular bundles. Radial walls of this endodermis have Casparian bands and tangential walls have relatively few plasmodesmata, so this cell layer restricts apoplasmic and symplasmic flow of carbon into nodules and nitrogen out of nodules.
The transport of solutes, including C and N metabolites but also metal ions, e.g. Fe and Mo required for nitrogenase, are exchanged via both plant and bacterial transporters on the symbiosome and bacterial membrane, and this transport is carefully controlled to balance supply and demand (Figure 4.49; Udvardi and Poole 2013). Not all rhizobia that invade and inhabit legume nodules fix nitrogen efficiently, with the legume host able to restrict C flow to those nodules that do not fix sufficient N.
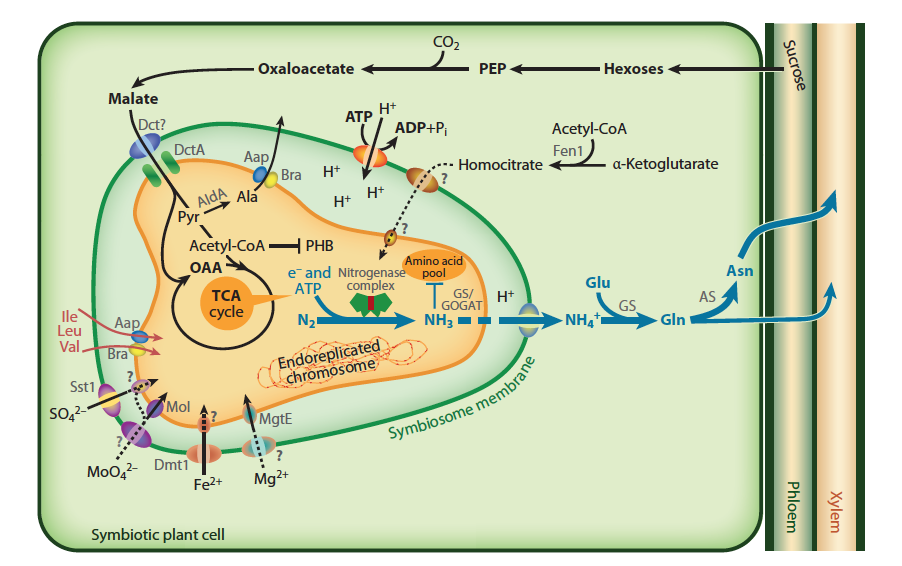
Figure 4.49 Transport and metabolism in an infected nodule cell. Sucrose from the shoot is converted to malate in the plant and imported across the symbiosome membrane and into bacteroids, where it fuels nitrogen fixation. The product of the nitrogen fixation is then exported back to the plant, where it is assimilated into asparagine (Asn) for export to the shoot (blue arrows). In many legumes, such as soybean, the export products are ureides instead of Asn. The plant must provide metals and ions to the bacteroid, although only some of the transport systems on the symbiosome and bacteroid membranes are defined. Many rhizobia lack the ability to make homocitrate or become symbiotic auxotrophs for supply of branched-chain amino acids and become dependent on the plant. (Reproduced with permission by Annual Reviews from M. Udvardi and P.S. Poole, Annu Rev Plant Biol 64: 781-805, 2013)
The concentration of nitrogenous solutes in the xylem apoplasm causes a hydrostatic pressure to develop, and this results in a mass flow of nodule xylem sap to adjacent roots. The water that accompanies sucrose entering the nodule as phloem sap is re-exported with assimilated nitrogen in the xylem. Nodules are thus analogous to ‘glands’ that secrete nitrogenous compounds.
Rates of N2 fixation can be measured by a number of techniques to address questions of nodule efficiency and nitrogen cycling in agricultural and natural plant systems. Nitrogenase is pivotal for initial reduction of N2 but this same enzyme will also reduce acetylene (C2H2) to ethylene (C2H4). Acetylene is an effective competitor with N2 for nitrogenase so the rate of C2H4 synthesis is proportional to nitrogenase activity. Acetylene reduction gives an instantaneous estimate of the N2 fixation rate. Another instantaneous technique requires flushing nodulated roots with an argon : oxygen gas mixture (79:21) to displace all N2. All electron flux through nitrogenase is then diverted to the reduction of protons to H2 rather than N2 to NH4+ (Equation 2). The rate of H2 evolution by roots can thus be used to estimate nitrogenase activity.
Alternative approaches to ‘instantaneous’ estimates of N2 fixation provide an integrated rate of fixation over periods of hours or days. The proportions of inorganic and organic nitrogen compounds in xylem sap are affected by the ratio of inorganic nitrogen taken up to symbiotic N2 fixation; this can be exploited in genera of legumes in which amides and ureides are major products of N2 fixation. Soybean, for example, exports less than 10% of nitrogen to shoots in the form of ureides when supplied nitrate but more than 80% when all nitrogen is biologically fixed. Thus, relative ureide levels in sap give an estimate of N2 fixation.
Many experiments now rely on 15N-based techniques to obtain an integral of fixation over the life of a plant. These techniques rely on a difference in ratio of the stable isotopes of nitrogen (15N and 14N) in soil and atmosphere (Figure 4.50). The soil must be enriched in 15N relative to the atmosphere — either naturally (the process of denitrification causes a fractionation of the two isotopes, leaving the soil enriched in 15N) or by artificial 15N addition. The N2-fixing plant of interest is sampled, together with an adjacent non-N2-fixing plant (e.g. grass) whose 15N enrichment represents that of soil nitrogen. 15N enrichment in digested plant material and soil is analysed isotopically in a mass spectrometer and contribution of biological N2 fixation calculated.
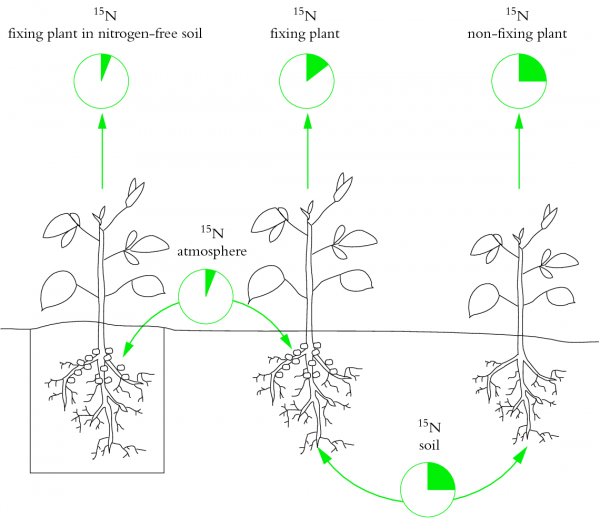
Figure 4.50 Basis of the natural abundance method for assessing the contribution of N2 fixation to legume nutrition. This method entails measuring plant 15N/14N ratio by mass spectrometry. Natural differences in 15N/14N ratio between soil and atmospheric nitrogen are exploited. Legumes to the left and right of the figure each have a unique source of nitrogen, while a test plant in the middle relies on both fixed nitrogen and soil inorganic nitrogen. Plants (left) denied a source of inorganic nitrogen (e.g. nitrate) fix atmospheric nitrogen and therefore have low 15N/14N ratios. Plants without nodules (right) take up only soil-derived nitrogen and are enriched with 15N (high 15N/14N ratios). 15N 'signatures' of these two sets of plants can be used to estimate the relative contributions of soil and atmospheric nitrogen as nitrogen sources in the test plant, and therefore to assess the significance of N2 fixation. (Based on Peoples et al. 1989; reproduced with permission of ACIAR)
A typical ‘good’ rate of fixation for a (non-irrigated) field of subtropical legumes in northern Australia is c. 60–100 kg N ha–1 year–1. About the same amount of nitrogen is harvested as seed from a crop of cowpea, soybean or chickpea, so growing these legumes does not add net nitrogen to the soil; it does, however, spare nitrogen which would otherwise be removed at harvest. Irrigated legume-based pastures in temperate Australia or New Zealand fix 250–300 kg N ha–1 year–1 and make a substantial contribution to the low energy costs of agriculture in these regions. Selection of appropriate biological N2 fixers could greatly improve N2 fixation in tropical legume crops.