Figp2.01.png
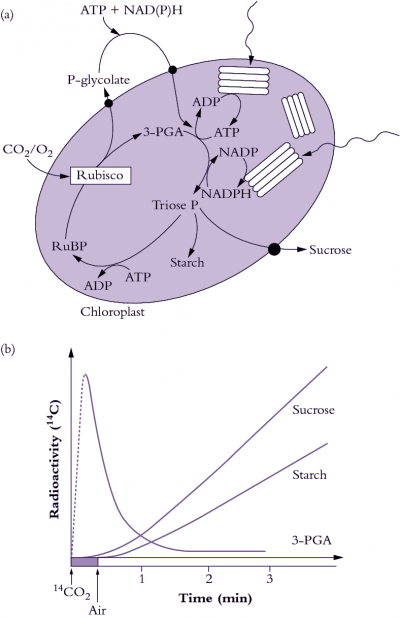
Figure 2.1 Photosynthetic carbon reduction (PCR cycle, also termed the Calvin-Benson cycle) utilises ATP and NADPH produced by thylakoid electron transport to drive CO2 fixation by Rubisco (a). CO2 is incorporated into a 5-carbon sugar phosphate to produce two 3-carbon sugar phosphates which can either be exported from the chloroplast for sucrose synthesis, be recycled to make more 5-carbon acceptors, or be used to make starch. The appearance of radioactive carbon in 3-carbon sugar phosphates and then in starch and sucrose following photosynthesis in 14CO2 was evidence for the pathway of photosynthesis. (b) (Original drawing courtesy Robert Furbank).
The biochemical pathway of CO2 fixation was discovered by feeding radioactively labelled CO2 in the light to algae and then extracting the cells and examining which compounds accumulated radioactivity. Figure 2.1(b) shows a typical labelling ‘pattern’ for a C3 plant. Here, a short burst of labelled CO2 was given to the plants, then the label was ‘chased’ through the photosynthetic pathway by flushing with unlabelled air. Atmospheric CO2 is initially incorporated into a five-carbon sugar phosphate (ribulose-1,5-bisphosphate or RuBP) to produce two molecules of the phosphorylated three-carbon compound 3-phosphoglycerate, often referred to as the acidic form 3-phosphoglyceric acid (3-PGA). Hence, plants which use Rubisco as their primary enzyme of CO2 fixation from the air are called C3 plants. Consequently, in C3 plants, 3-PGA is the first labelled sugar phosphate detected after a pulse of 14CO2 has been supplied (Figure 2.1b). In the PCR cycle, 3-PGA is phosphorylated by the ATP produced from thylakoid electron transport (see Chapter 1) and then reduced by NADPH to produce triose phosphate. Triose phosphates are the carbon backbones, produced by the PCR cycle, for the synthesis of critical carbohydrate for the maintenance of plant growth and the productive yield of stored carbohydrate in seed.
Newly synthesised triose phosphate faces three options. It can be (1) exported to the cytosol for sucrose synthesis and subsequent translocation to the rest of the plant, (2) recycled within the chloroplast to produce more RuBP or (3) diverted to produce starch (Figure 2.1a). This is shown by the time-course of the appearance of radioactivity in starch and sucrose after it has passed through 3-PGA (Figure 2.1b). The energy requirements of the PCR cycle are three ATP and two NADPH per CO2 fixed, in the absence of any other energy-consuming processes.
Sucrose and starch synthesis
Most of the triose phosphate synthesised in chloroplasts is converted to either sucrose or starch. Starch accumulates in chloroplasts, but sucrose is synthesised in the surrounding cytosol, starting with the export of dihydroxyacetone phosphate and glyceraldehyde phosphate from the chloroplast. A condensation reaction, catalysed by aldolase, generates fructose-1,6-bisphosphate, and this is converted to fructose-6-phosphate after an hydrolysis reaction catalysed by fructose-1,6-phosphatase. Sucrose-6-phosphate synthase then generates sucrose-6-phosphate from the reaction of fructose-6-phosphate and UDP-glucose. The phosphate group is removed by the action of sucrose-6-phosphatase. This Pi is transported back into the chloroplast where it is available for ATP synthesis. For each molecule of triose phosphate exported from a chloroplast, one Pi is translocated inwards.
Sucrose synthesised within the cytosol of photosynthesising cells is then available for general distribution and is commonly translocated to other carbon-demanding centres via the phloem (see Chapter 5).
By contrast, starch synthesis occurs within chloroplasts. The first step is a condensation of glucose-1-phosphate with ATP. Starch synthase then transfers glucose residues from this molecule to the non-reducing end of a pre-existing molecule of starch. Starch consists of two types of glucose polymer, namely amylose and amylopectin. Amylose is a long, unbranched chain of D-glucose units connected via (α1–4) linkages. Amylopectin is a branched form, with (α1–6) linkages forming branches approximately every 24–30 glucose residues.
