4.2-Ch-Fig-4.18.png
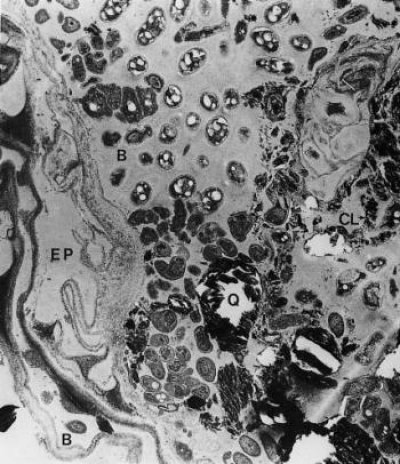
Figure 4.18 Mature rhizosphere from roots of clover (Trifolium subterraneum L.). The outer cortex has been crushed and epidermal cells (EP) have become distorted, leading to leakage of substrates into the rhizosphere. The rhizosphere is rich in microorganisms with bacteria (B) clearly visible. Soil (Q) and clay (CL) particles are held together in the inner rhizosphere by a mucilage of polysaccharides. Sustained losses of carbon required to maintain this microflora are thought to come from exudation and senescence of root cells. (× 10,000) (Courtesy R.C. Foster, A.D. Rovira and T.W. Cock)
Microbial activity, sustained by photoassimilates secreted from roots, contributes substantially to rhizosphere properties. The level of microbial activity is also influenced by availability of nitrogen as a substrate for microbial growth. Soils with high fertility and biological activity have microbial densities 5–50 times greater in the rhizosphere than in bulk soil. The diversity of rhizosphere microflora is spectacular (Figure 4.18) and still incompletely described. An initial hurdle in the identification of rhizosphere microbes was the fact that most of them are unculturable on most known growth media. Metagenomics allows species to be sequenced from soil samples without culturing, largely overcoming this bottleneck. Next Generation sequencing studies of microbes present on root surfaces and in the rhizosphere soil have discovered thousands of different bacterial and fungal species living in close association with plant roots (e.g. Bakker et al. 2013; Bulgarelli et al. 2012; Lundberg et al. 2012). Some of these are very abundant and found in association with many plant species, others are less abundant and highly variable (Figure 4.19).
4.2-Ch-Fig-4.19.png
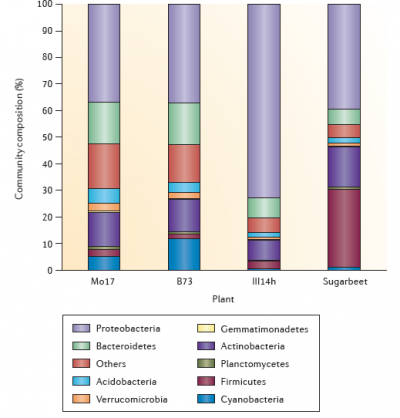
Figure 4.19 The composition of the bacterial community in the rhizosphere. The figure shows examples of the composition of the bacterial community in the rhizosphere of three maize genotypes (Mo17, B73 and III14h) and of sugarbeet. The distribution of the different bacterial phyla is based on data obtained by 454 sequencing (maize) and G3 PhyloChip analyses (sugarbeet). The bacterial community composition was characterized in the rhizosphere of 27 maize genotypes cultivated in five fields located in three states in the USA. Here, three genotypes displaying contrasted rhizosphere microbiota in a given field are depicted for illustration and the sugar beet rhizosphere microbiota presented is from seedlings grown in a disease-conducive soil in The Netherlands. (Reproduced by permission from Macmillan Publishers Ltd from L. Philippot et al. Nature Rev Microbiol 11: 789-799, 2013)
These microbes can almost be viewed as an extension of the plant into the soil. Like the human gut microbiome, the plant rhizosphere microbiome appears to be an essential part of the plant with multiple functions in nutrition and pathogen defense; it is inseparable from the plant and has been dubbed the plants second genome. The rhizosphere community is highly structured and not a random collection of species – it is strongly influenced by plant species and even ecotypes, by the type of the soil, availability of nutrients and the exudation of chemicals from the root (Bakker et al 2013). Plant mutants with altered chemical composition of root exudates have been found to attract significantly altered microbial communities. It will be fascinating to discover to what extent this is an active strategy of the plant to attract the most appropriate rhizosphere microbiome to help the plant survive in a given environment.
Rhizosphere microorganisms are also not uniformly distributed along roots. Apices are almost free of microbes but densities can increase dramatically in subapical zones. Very mature root axes with lateral branches are sparsely populated with microbes. Even within these zones, there are large variations in distribution, with radial epidermal walls of roots secreting exudates which can support huge microbial populations, up to 2 × 1011 microbes cm–3. Composition of microbial communities varies with their distribution along the root as well, likely reflecting different nutrient sources along the root. Fluorescence in situ hybridization (FISH) can be used to visualise different taxa of bacteria on the root surface (rhizoplane; Figure 4.20).
4.2-Ch-Fig-4.20.png
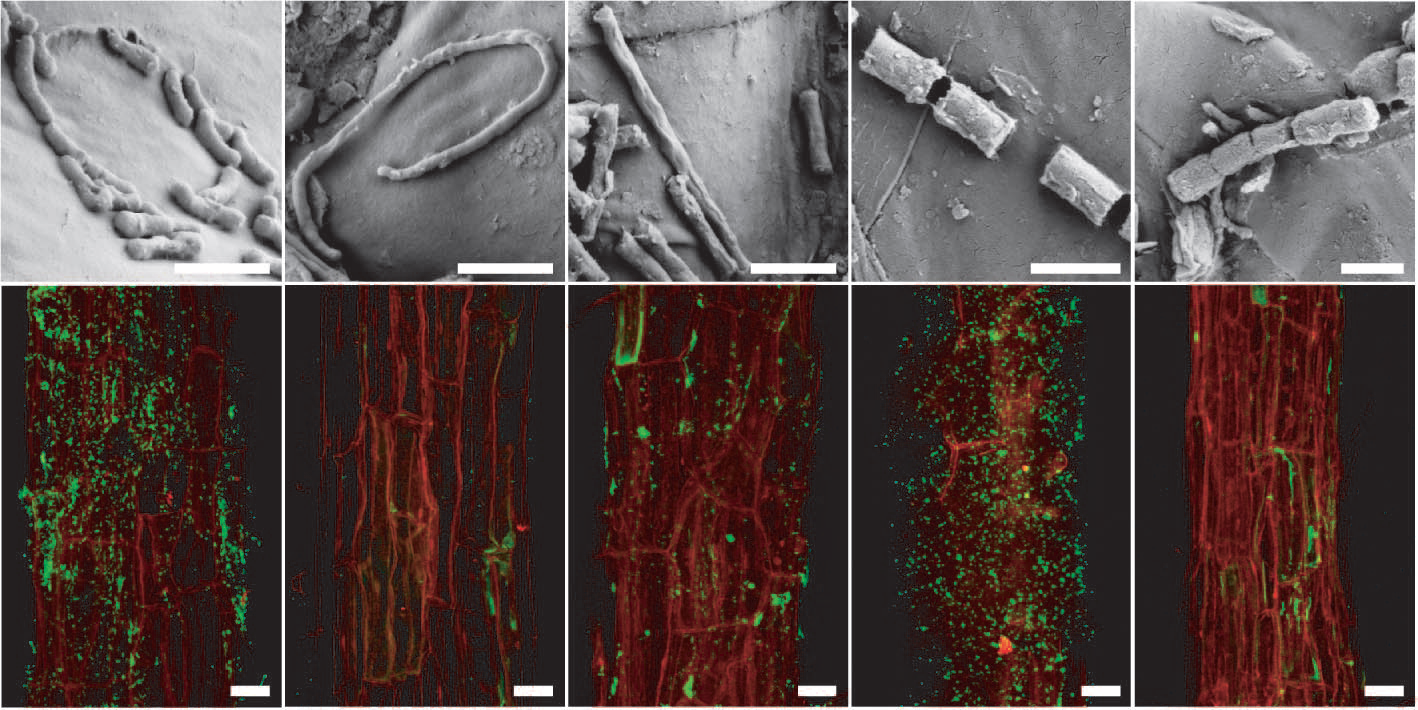
Figure 4.20 Arabidopsis root-inhabiting bacteria are detectable on the rhizoplane. a to e, Scanning electron micrographs of bacteria-like structures. Bars, 1 mm. f to j, Detection of bacteria by fluorescence in situ hydridisation (FISH) using probes against specific bacterial groups (bacteria in green due to AlexaFluor488) on the root surface (red, root autofluorescence) by confocal laser scanning microscopy. f, Most Eubacteria detected with probe EUB338. g, Negative control with reverse complementary probe of EUB338 (NONEUB). h, Betaproteobacteria detected with probe BET42a. i, Bacteroidetes detected with probe CF319a. j, Actinobacteria detected with probe HGC69a. Bars, 20 mm. (Reproduced by permission from Macmillan Publishers Ltd from D. Bulgarelli D et al., Nature 488: 91-95, 2012)
Roots do of course influence adjacent soil throughout their length by setting up gradients of water, gases and ions. For example, in waterlogged soils leakage of O2 from aerenchymatous roots leads to oxidation of metal ions and local build up of aerobic microflora around roots of agricultural plants (Chapter 18). In general, however, the most active microbial populations and rates of chemical transformation in the rhizosphere occur in the subapical zones of the root. In supporting these processes, root-associated microbes metabolise inorganic nitrogen, depositing protein nitrogen in the process of immobilisation. Microbial activity also produces plant growth regulators such as auxin, cytokinins and gibberellins, sometimes in amounts sufficient to influence root morphogenesis. Ethylene can also be produced by rhizospheric fungi, potentially influencing root morphological changes such as lateral root initiation. Some bacteria have been found to promote plant growth by reducing ethylene levels around roots through production of an enzyme degrading an ethylene precursor, 1-aminocyclopropane-1-carboxylate (ACC) deaminase.
